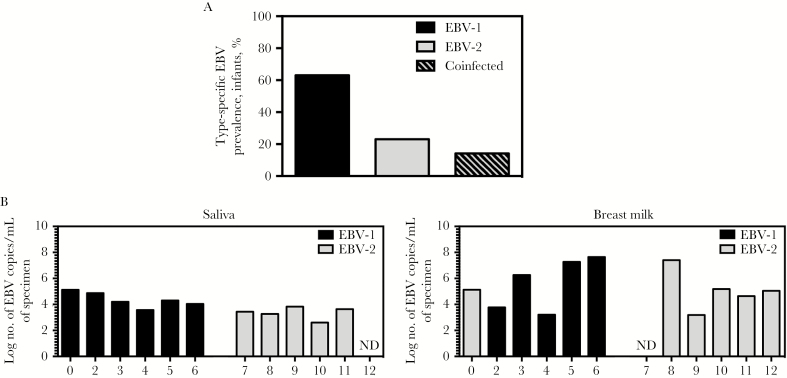
Figure1.
Prevalence of Epstein-Barr virus (EBV) types in the Kisumu, Kenya, infant population and EBV types detectable in saliva and breast milk specimens from the infants’ mothers. A, Blood specimens were collected from infant donors at 6 weeks, 10 weeks, 14 weeks, 18 weeks, 6 months, 9 months, and 12 months of age. DNA was isolated from blood specimens and used to perform quantitative polymerase chain reaction (qPCR) analysis to detect BALF5 in the EBV genome (and thereby identify EBV-positive samples (84 collected at age 6 weeks, 62 collected at 10 weeks, 37 collected at 14 weeks, 23 collected at 18 weeks, 28 collected at 6 months, 28 collected at 9 months, and 32 collected at 12 months). DNA samples that were positive for EBV genome were further analyzed to determine the EBV type present in each sample, via a multiplex reverse-transcription PCR for EBNA3c that distinguishes EBV type 1 (EBV-1) and EBV-2; for 3 children, we determined the EBV type for samples collected ≥1 time point. DNA samples that were positive for EBV-1 or EBV-2 through the first year of life were used to determine the type-specific EBV prevalence in this population. Infants were considered coinfected if both EBV-1 and EBV-2 were detected at any time point tested. Prevalence data for samples in which the EBV type could be determined are shown in bar graph (22 of 35 were positive for EBV-1, 8 of 35 were positive for EBV-2, and 5 of 35 were positive for both types). B, Saliva and breast milk specimens were collected from infants’ mothers at postpartum week 6, and DNA was isolated. Multiplex quantitative PCR analysis of BALF5 in the EBV genome was performed to determine the log number of EBV copies per milliliter of saliva and breast milk. Additionally, multiplex reverse-transcription PCR analysis to detect EBNA3c was performed to identify the EBV type present in each sample. Abbreviation: ND, not detected.