Retardation of growth induced by water stress is attenuated by the overexpression of AtPgb1 and aggravated when it is suppressed. Expression of AtPgb1 is required to delay the death of root tips and to prevent the premature differentiation of meristematic cells.
Keywords: Arabidopsis roots, auxin, polyethylene glycol, phytoglobin, root meristem, water stress
Abstract
Maintenance of a functional root is fundamental to plant survival in response to some abiotic stresses, such as water deficit. In this study, we found that overexpression of Arabidopsis class 1 phytoglobin (AtPgb1) alleviated the growth retardation of polyethylene glycol (PEG)-induced water stress by reducing programmed cell death (PCD) associated with protein folding in the endoplasmic reticulum (ER). This was in contrast to PEG-stressed roots down-regulating AtPgb1 that exhibited extensive PCD and reduced expression of several attenuators of ER stress, including BAX Inhibitor-1 (BI-1). The death program experienced by the suppression of AtPgb1 in stressed roots was mediated by reactive oxygen species (ROS) and ethylene. Suppression of ROS synthesis or ethylene perception reduced PCD and partially restored root growth. The PEG-induced cessation of root growth was preceded by structural changes in the root apical meristem (RAM), including the loss of cell and tissue specification, possibly as a result of alterations in PIN1- and PIN4-mediated auxin accumulation at the root pole. These events were attenuated by the overexpression of AtPgb1 and aggravated when AtPgb1 was suppressed. Specifically, suppression of AtPgb1 compromised the functionality of the WOX5-expressing quiescent cells (QCs), leading to the early and premature differentiation of the adjacent columella stem cells and to a rapid reduction in meristem size. The expression and localization of other root domain markers, such as SCARECROW (SCR), which demarks the endodermis and QCs, and WEREWOLF (WER), which specifies the lateral root cap, were also most affected in PEG-treated roots with suppressed AtPgb1. Collectively, the results demonstrate that AtPgb1 exercises a protective role in roots exposed to lethal levels of PEG, and suggest a novel function of this gene in ensuring meristem functionality through the retention of cell fate specification.
Introduction
Among several types of abiotic stress, drought is the most adverse environmental factor affecting plant growth and limiting crop productivity (Westgate and Boyer, 1985). Conditions of water deficit influence both root and shoot behaviour, and plants have developed complex and diverse tolerance and avoidance strategies that are often genotype-specific. While tolerance strategies are usually responses that allow plants to cope with and tolerate the adverse environmental conditions, avoidance strategies allow plants to ‘circumvent’ the stress (Chaves et al., 2002). Both strategies often require extensive reprogramming of gene expression accompanied by metabolic and structural changes (Bohnert and Sheveleva, 1998). A rapid increase in stomatal resistance and alterations in gas exchange in response to growth regulators produced by dehydrating roots are early strategies to limit water loss in photosynthetic tissue (Chaves et al., 2002). These events are often followed by a reduced root growth rate, due to imbalanced cellular water potential compromising elongation, and mechanical impedance from inefficient root penetration in water-depleted soils (Bengough et al., 2011).
In roots, the iterative generation of cells is ensured by the activity of the root apical meristem (RAM), which surrounds a group of mitotically inactive quiescent cells (QCs). The QCs act as an ‘organizing centre’, with the main function to maintain the adjacent stem cells in an undifferentiated state. Stem cells located at the proximal and lateral side of the RAM contribute to the formation of the vascular tissue, endodermis, and cortex, while those situated distally to the QCs are the progenitors of the central domain of the root cap, the columella (Dolan et al., 1993) (see Supplementary Fig. S1A at JXB online). This precise and conserved organizational pattern, best exemplified in Arabidopsis, is regulated by a complex genetic network including the transcription factors WUSCHEL RELATED HOMEOBOX 5 (WOX5), SCARECROW (SCR), and WEREWOLF (WER) (Petricka et al., 2012). Alteration of one or more of these factors, expressed in specific root domains (Supplementary Fig. S1B), perturbs the exquisitely regulated regenerative nature of the RAM, resulting in retardation or cessation of growth. A well-documented example is the loss of the undifferentiated state of the columella stem cells following experimental removal of WOX5 expression (Pi et al., 2015).
Cell fate specification and maintenance in the RAM is influenced by the PIN-mediated transport of auxin which, through a basipetal flow, accumulates in the root tip. Among the several PIN members, the most well characterized in relation to root behaviour are PIN1, which with its basal localization on the plasma membrane creates a root tip-directed auxin flow (Vieten et al., 2005), and PIN4, which is implicated in the short-range redistribution of auxin within the RAM (Friml et al., 2002) (see Supplementary Fig. S1A). Pharmacological treatments or genetic manipulations that alter either the flow of auxin or the establishment of auxin maxima at the tip disrupt the patterning and functionality of the RAM, hence compromising root growth.
Perturbations in RAM function occur under conditions of water deficit, with a mild stress causing the premature differentiation of the RAM (Ji et al., 2014), while a more severe stress triggers the death of the meristematic cells (Duan et al., 2010). Autophagic programmed cell death (PCD) of the meristematic cells has been implicated in root-tip death of several species, including pea and maize exposed to severe stress conditions (Subbaiah and Sachs, 2003). Widespread death was also observed in Arabidopsis RAMs grown in a polyethylene glycol (PEG) environment generating water potentials less than –1.40 MPa (Duan et al., 2010). The death program was triggered by an imbalance between folded and unfolded proteins in the endoplasmic reticulum (ER), which represents a common cellular stress (Williams et al., 2014). In both animals and plants, adverse environmental conditions contribute to the accumulation of misfolded proteins in the secretory pathway which, if not mitigated by the activation of the highly conserved unfolded protein response (UPR), leads to apoptosis and PCD (Boyce and Yuan, 2006). Attenuators of the ER-stress mediated death program, such as BAX inhibitor-1 (BI-1), reduce cellular death (Watanabe and Lam, 2006).
Retention of meristem function and protection of meristematic cells from death under suboptimal environmental conditions have been postulated to be an avoidance strategy modulated by phytoglobins (Pgbs) (Mira et al., 2017). Phytoglobins, formerly known as non-symbiotic plant hemoglobins (Hill et al., 2016), are heme-containing proteins that act as scavengers of nitric oxide (NO) and suppressors of the death program (Huang et al., 2014). Expressed in the root tip (Dordas et al., 2003; Zhao et al., 2008) and induced under conditions of biotic and abiotic stress, Pgbs exercise a protective role during hypoxic (low-oxygen) conditions (Hill, 2012). Overexpression of Pgbs enhanced survival of hypoxic roots in Arabidopsis (Hunt et al., 2002), alfalfa (Dordas et al., 2003), and maize (Mira et al., 2016b). In maize, the overexpression of Pgbs contributes to the retention of a functional meristem by repressing the death program triggered by oxygen deprivation (Mira et al., 2016b). Based on these premises, we speculated that the protective function of Pgbs represents a strategy adopted by plants to cope with diverse types of stress, including severe water deficit. This notion was tested in the present study by evaluating the effects of altered expression of the Arabidopsis class I Pgb (AtPgb1) on the behaviour and structure of the RAM in roots experiencing severe PEG-induced water stress that caused death of wild-type root cells.
Material and methods
Plant material
Arabidopsis thaliana (Columbia) plants overexpressing (35S:Pgb1) or suppressing (Pgb1-RNAi) phytoglobin1 were those characterized by Hebelstrup et al. (2006). The β-glucuronidase (GUS) or green fluorescent protein (GFP) reporter lines crossed with the 35S:Pgb1 and Pgb1-RNAi lines included PIN1-GFP (Fernandez-Marcos et al., 2011), WOX5:GFP and SCR:GFP (Zhang et al., 2015), WER:GFP (Rodriguez et al., 2015), PIN4:GUS and DR5:GUS (Li et al., 2015), CYCB1;2:GUS (Zhang et al., 2010a), and CYCB1;3:GUS and CYCA1;2:GUS (Bulankova et al., 2013). Crossing was conducted (as in Mira et al., 2016c) using the 35S:Pgb1 and Pgb1-RNAi lines as pollen donors. Arabidopsis seeds were sterilized in 70% ethanol containing 0.5% Triton X-100 for 15 min followed by 95% ethanol for 15 min (Mira et al., 2016c) The seeds were then plated on agar germination medium [Murashige and Skoog (MS) salt with vitamins, 2.5% sucrose (w/v) and 0.8 % agar (w/v), pH 5.7], incubated for 24 h at 4 °C in the dark, and then transferred to a growth cabinet (22 °C, 16 h light/8 h dark). The PEG-containing medium was prepared as previously described by Duan et al. (2010). Briefly, the desired concentration of PEG-8000 was dissolved in full-strength MS salts containing 2.5% sucrose (pH 5.8). The solution was autoclaved [121 °C, 20 psi (~138 kPa)] for 10 min, cooled to room temperature, and poured on solid medium (full-strength MS salts, 2.5% sucrose and 0.8% agar). The plates were incubated at room temperature for 48 h to allow the PEG solution to infiltrate the solid medium. Arabidopsis seedlings (4 d-old) were placed on freshly prepared PEG medium and the growth of the root was measured at 0, 2, 4, and 6 d.
Chemical treatments
The inhibitor of ethylene perception 1-methylcyclopropene (1-MCP) was administered at a concentration of 1 ppm (Takahashi et al., 2015). The seedlings were incubated with 1-MCP in a sealed container during the PEG treatment. Diphenyleneiodonium chloride (DPI), an inhibitor of NADPH oxidase (Huang et al., 2014), and sodium 4-phenylbutyrate (PBA), an attenuator of ER-stress (Welch and Brown, 1996), were applied at concentrations of 40 μM and 1 mM respectively. The solutions (10 μl) were dispensed every other day directly on the PEG-treated roots. The ethylene-releasing agent Ethephon (ET) (Zhang et al., 2010b) was applied at a concentration of 10 μM.
Microscopy
Roots were stained with propodium iodide (10 µg ml–1) for 5 min, rinsed with distilled water, mounted with 10% glycerol, and visualized using a confocal Zeiss LSM 700 microscope. Signals for propodium iodide and GFP were visualized using a 570–670 nm filter and 500–550 nm filter, respectively. Image J (https://imagej.nih.gov/ij/index.html) was used to measure the GFP signal and the area of expression. The size of the meristem (calculated as described by González-García et al., 2011) included the region from the QCs to cells that had twice the length of the immediately preceding ones. Starch granules in the columella cells were visualized using the Lugol’s staining method (Hong et al., 2015). Roots were immersed in Lugol’s solution (Sigma) for 30 s, cleared with a solution containing chloral hydrate, glycerol, and water in a 8:3:1 ratio, and imaged by differential interference contrast microscopy. For GUS staining, roots were immersed in acetone for 10 min, rinsed twice in 100 mM sodium phosphate buffer (pH 7.2), infiltrated for 15 min with 100 mM sodium phosphate buffer (pH 7.2) containing 1 mg ml–1 5-bromo-4-chloro-3-indolyl β-glucuronide, 10 mM EDTA, 10 mM potassium ferrocyanide, and 0.1% Triton X-100, and incubated at 37 °C for 5 h (Mira et al., 2016c).
ROS and PCD detection
A terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay was performed to detect PCD using an In Situ Cell Death Detection Kit (Roche), according to the manufacturer’s instructions. Cell death was also estimated by staining the roots with Trypan blue and propidium iodide as described previously (Leite et al., 1999; Ning et al., 2002). Reactive oxygen species were localized using dihydroethidium, as reported by Mira et al. (2016b).
DNA isolation and electrophoresis
For DNA fragmentation analyses, 100 mg of the root tips were frozen and ground in liquid nitrogen. The samples were digested using cellulase (1%) for 2 h. DNA extraction was performed using the Apop-ladder EX™ DNA fragmentation assay kit (Clontech Laboratories Inc.) according to the manufacturer’s protocol. The samples were separated on 2% agarose gel.
Ethylene measurements
Ethylene measurements on Arabidopsis seedlings were performed as previously described (Mira et al., 2016b). Briefly, 200 mg fresh weight of seedlings was incubated in a sealed 3-ml syringe for 3 h in the dark at 22 °C. The gas (1 ml) accumulated in the headspace was analysed with a Bruker 450-GC Gas Chromatograph. Data analysis was carried out using the Bruker Compass Data analysis 3.0 software. All experiments were performed in triplicate.
Gene expression studies
Extraction of RNA from roots was performed using TRI Reagent (Mira et al., 2016c). The total RNA was treated with DNase I (recombinant, RNase-free, Roche). A High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) was used for cDNA synthesis. Quantitative RT-PCR was performed as previously described (Mira et al., 2016c). All primers used for gene expression studies are listed in Supplementary Table S1. The relative gene expression level was analysed using the 2−∆∆CT method (Livak and Schmittgen, 2001) with UbQ10 as the reference gene.
Statistical analysis
Data were analysed by one-way ANOVA using the SPSS program (IBM Corp. Released 2010. IBM SPSS Statistics for Windows, Version 19.0. Armonk, NY: IBM Corp.). Treatments means were compared using Duncan’s test (α=0.05) to differentiate the significance of differences between various parameters.
Results
Modulation of AtPgb1 expression influences root sensitivity to water stress
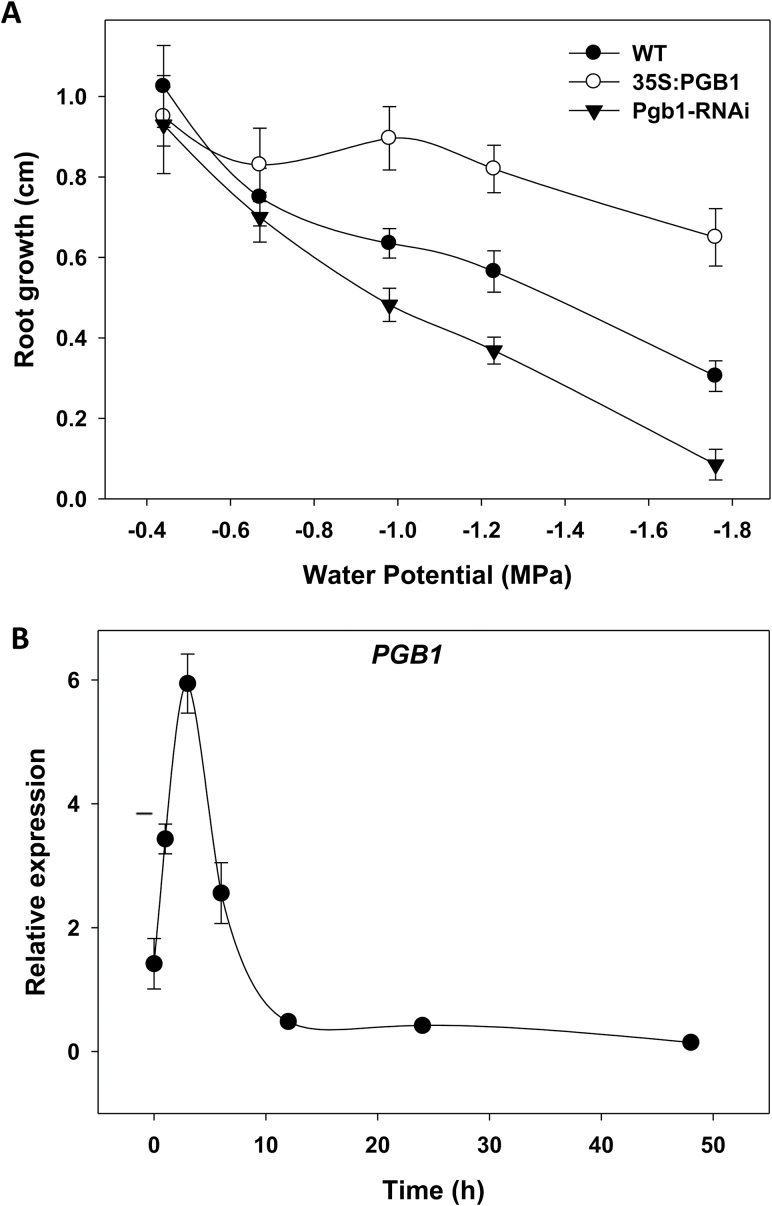
Drought stress in 4-d-old Arabidopsis seedlings was induced by infusing the medium with different concentrations of PEG-8000 corresponding to water potential values ranging from –0.42 MPa (0% PEG) to –1.76 MPa (40% PEG) (see Supplementary Fig. S2). Autoclaving the PEG solution for only 10 min had no significant effects on the water potential, as similar values were obtained when the PEG solution was filter-sterilized (Supplementary Fig. S2). In wild-type (WT) seedlings, elongation of the primary root was inhibited with increasing levels of PEG, which lowered the water potential of the medium (Fig. 1A). The inhibitory effect of PEG was more pronounced in Arabidopsis plants suppressing AtPgb1 (line Pgb1-RNAi), where root elongation was almost completely inhibited with 40% PEG (–1.76MPa). This was in contrast to plants overexpressing AtPgb1 (line 35S:Pgb1), which showed reduced sensitivity to water deficit and substantial root growth at the highest PEG levels (Fig. 1A and Supplementary Fig. S3). As the most significant differences in root growth patterns among lines were observed with 40% PEG (–1.76 MPa), this concentration was used for all the subsequent experiments. A time-course of root elongation of the three lines at –1.76 MPa is shown in Supplementary Fig. S4.
Fig. 1.
(A) Effects of decreasing water potential on root growth of a wild-type (WT) line and lines overexpressing (35S:Pgb1) or down-regulating (Pgb1-RNAi) AtPgb1. Arabidopsis seedlings were 4 d old when placed on PEG-containing medium, and root length was measured after 6 d. Values are means ±SE of three biological replicates. (B) Time-course expression of AtPgb1in wild-type roots grown in 40% PEG (–1.76 MPa). Expression values are means ±SE of three biological replicates.
The expression of AtPgb1 was monitored in WT roots cultured at –1.76 MPa (Fig. 1B). Water stress induced a rapid increase in the transcript levels of AtPgb1, which reached a peak after 3 h in PEG before declining. An increase in AtPgb1 expression, albeit less pronounced, was also observed on lower levels of PEG (see Supplementary Fig. S5).
AtPgb1 suppresses ER stress-mediated PCD in PEG-treated roots
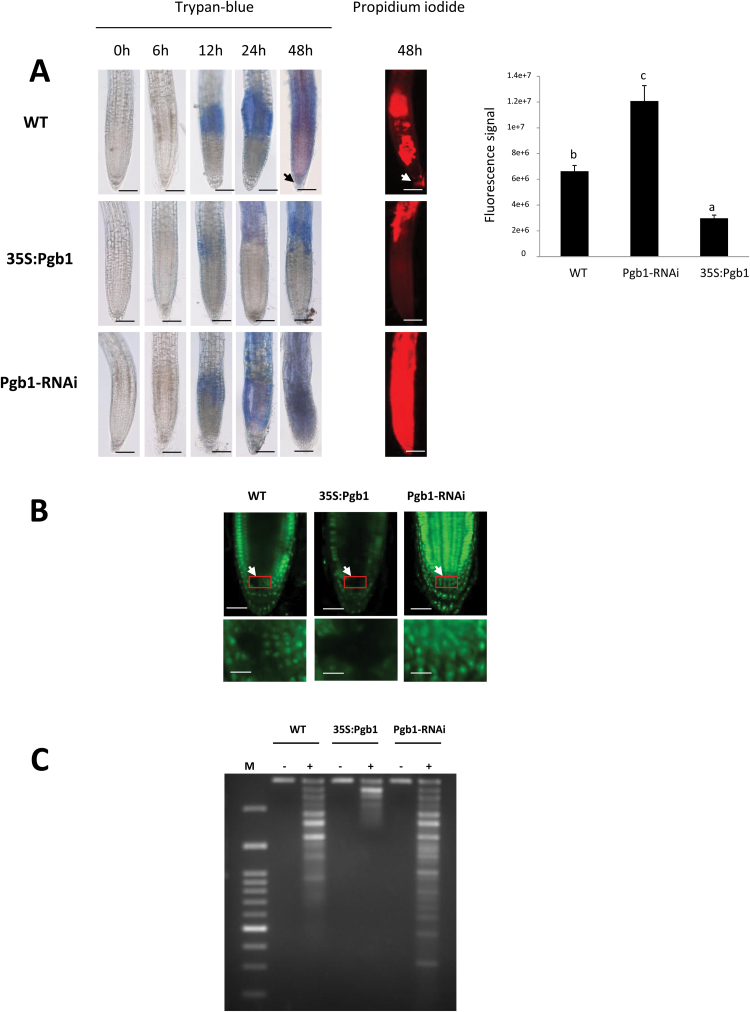
Many types of stress, including water deficit, limit plant growth by inducing death in root cells (Subbaiah and Sachs, 2003; Duan et al., 2010). Cell death progression in Arabidopsis roots subjected to water stress was examined using Trypan blue, a dye that can only penetrate damaged plasma-membranes and stain dead cells (Huang et al., 2014). In WT roots, substantial Trypan staining was initially apparent after 12 h of water deficit within the upper domain of the roots encompassing the elongation and differentiation zones (Fig. 2A). Following prolonged PEG treatment, the signal progressed apically along the root profile, reaching the RAM at 48 h (arrow in Fig. 2A). Overexpression of AtPgb1 (line 35S:Pgb1) delayed cell death, which was excluded from the root tip, while roots suppressing AtPgb1 (line Pgb1-RNAi) were almost completely stained by Trypan blue after only 24 h of PEG treatment (Fig. 2A). The staining pattern of Trypan blue at 48 h in PEG was also confirmed using propidium iodide, another stain unable to penetrate live cells (Ning et al., 2002) (Fig. 2A).
Fig. 2.
Programmed cell death (PCD) in roots exposed to 40% PEG (–1.76 MPa). (A) Trypan blue and propidium iodide staining of roots of the wild-type (WT) and lines overexpressing (35S:Pgb1) or down-regulating (Pgb1-RNAi) AtPgb1. Roots were grown on 40% PEG and stained at different time points (as indicated). The arrow indicates staining in the root tip of WT roots. The graph indicates the fluorescence signal (in pixels) of propidium iodide within the root apical tip (1 mm). Scale bars are 100 μm. Values are means ±SE of three biological replicates. Different letters on the graph indicate statistically significant differences (P<0.05). (B) Terminal deoxynucleotidyl transferase dUTP nick-and labelling (TUNEL) staining in the root tips after 48 h in 40% PEG. Arrows indicate the centre of the root apical meristem (RAM). The bottom panels are higher magnification images of the RAMs enclosed in the red boxes. Scale bars are 50 μm (upper panels) and 20 μm (lower panels). (C) Fragmentation of DNA extracted from roots of different lines grown for 48 h in the presence (+) or absence (–) of PEG. M, molecular weight marker.
As documented previously (Duan et al., 2010), the death program induced in root cells by water deficit is a type of PCD. This was apparent in the present study after 48 h in PEG. TUNEL staining was detected in several nuclei along the profile of WT roots, and in close proximity to the RAM (arrow in Fig. 2B). This was in contrast to roots up-regulating AtPgb1 where the death program was restricted to very few cells, and excluded the RAM. Many TUNEL-positive nuclei were detected in Pgb1-RNAi roots (Fig. 2B). This trend was also confirmed by the DNA profile, which revealed some laddering in WT and AtPgb1-suppressing roots, but not in roots where the level of AtPgb1 was increased (Fig. 2C). Detection of only a moderate DNA ladder, as observed in our study, has been ascribed to the fact that during water stress not all of the root cells undergo PCD (Gunawardena et al., 2004).
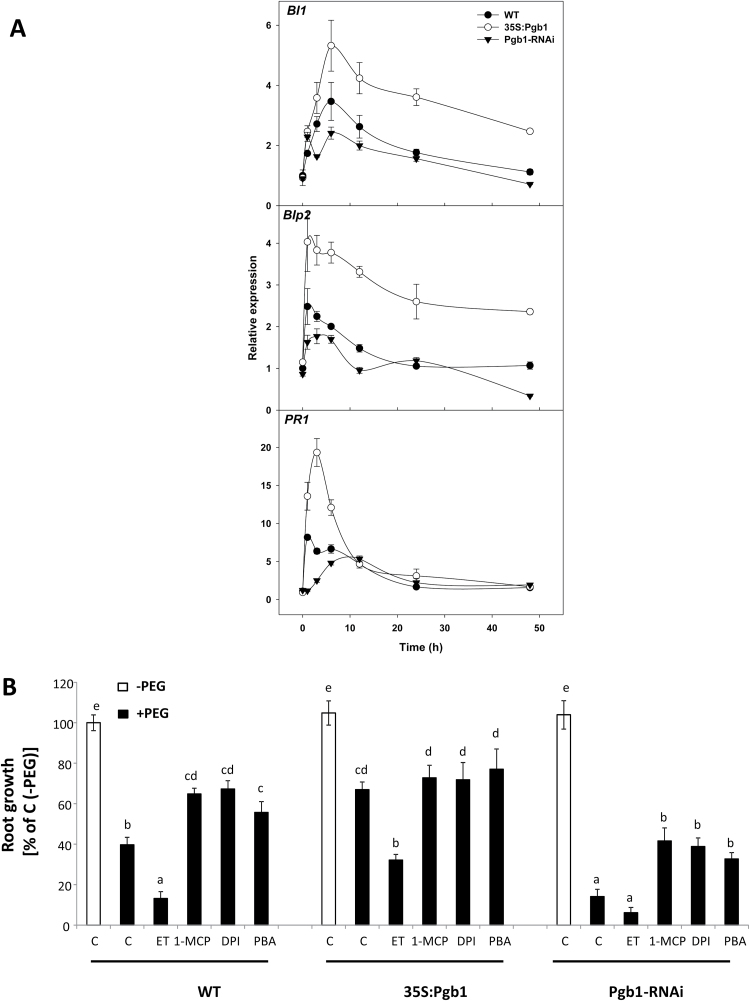
The ER participates in PCD by sensing and relaying cellular apoptotic signals through the balance between folded and unfolded proteins (Boyce and Yuan, 2006). Over-accumulation of misfolded or unfolded proteins is perceived as a form of stress which, if not mitigated by the activation of the unfolded protein response (UPR), escalates to PCD (Boyce and Yuan, 2006). The expression levels of the attenuator of the ER stress-mediated PCD, BAX Inhibitor-1 (BI-1) (Watanabe and Lam, 2008), and the markers of UPR activation, Luminal binding protein 2 (BiP2) and Pathogenesis-related protein 1 (PR1) (Duan et al., 2010), were highly induced, especially within the first 10 h, in water-stressed roots overexpressing AtPgb1 (Fig. 3A). This was in contrast to roots suppressing AtPgb1 which, relative to the WT, exhibited the lowest expression levels of the three genes (Fig. 3A). These transcriptional studies suggest that expression of AtPgb1 attenuates ER stress.
Fig. 3.
Endoplasmic reticulum (ER) stress-induced PCD in water-stressed roots. (A) Expression levels of BAX Inhibitor-1 (BI-1), Luminal binding protein 2 (BiP2), and Pathogenesis-related protein 1 (PR1) in roots treated with 40% PEG (–1.76 MPa) for the wildtype (WT) and lines overexpressing (35S:Pgb1) or down-regulating (Pgb1-RNAi) AtPgb1. Values are normalized to the WT at 0 h (set at 1), and are means ±SE of three biological replicates. (B) Effects of 4-phenyl butyric acid (PBA), 1-methylcyclopropene (1-MCP), ethephon (ET), and diphenyleneiodonium (DPI) on root growth. Values are expressed as percentages relative to the respective control in the absence of PEG (C, –PEG) (set at 100%), and are means ±SE of three biological replicates, each consisting of 20 roots. Different letters indicate statistically significant differences (P<0.05). When applied, PEG was added at a concentration of 40% (–1.76 MPa).
To further elucidate the role of AtPgb1 in alleviating ER stress-mediated PCD we used 4-phenyl butyric acid (PBA), a stabilizer of protein confirmation and attenuator of ER stress (Welch and Brown, 1996). Applications of PBA to the AtPgb1-suppressing roots partially relieved the PEG-growth inhibition (Fig. 3B), possibly by suppressing PCD (see Supplementary Fig. S6).
Collectively, these findings confirm the notion that severe water deficit induces PCD in root apices (Duan et al., 2010), and suggest that modulation of AtPgb1 expression influences root growth by regulating ER stress-mediated PCD of meristematic cells. Relative to the WT, growth inhibition and PCD were induced in roots where AtPgb1 was down-regulated and mitigated in those where AtPgb1 was up-regulated.
AtPgb1 regulation of death in PEG-stressed roots is mediated by ethylene and ROS
We have previously shown that the protective role of Pgbs on hypoxic maize root cells is exercised through the suppression of ethylene and ROS, elicitors of PCD (Mira et al., 2016a, 2016b). To establish if similar mechanisms operate during drought, we conducted studies on ethylene and ROS in PEG-treated roots. Relative to the WT, ethylene accumulation (measured in whole seedlings due to limitations in harvesting sufficient root tissue) was more pronounced in water-stressed seedlings suppressing AtPgb1 and less pronounced in those where the level of AtPgb1 was up-regulated (Table 1). These changes were most likely the result of transcriptional changes of key ethylene biosynthetic genes (see Supplementary Fig. S7).
Table 1.
Ethylene levels (nmol g–1 FW h–1) measured in Arabidopsis seedlings of the wild-type (WT) and lines overexpressing (35S:Pgb1) or down-regulating (Pgb1-RNAi) AtPgb1 cultured for 24 h in 40% PEG (–1.76 MPa). Values are means ±SE of three biological replicates. Different letters indicate statistically significant differences (P<0.05)
| –PEG | +PEG | |
|---|---|---|
| WT | 0.135 ± 0.016a | 0.422 ± 0.048c |
| 35S:Pgb1 | 0.137 ± 0.014a | 0.292 ± 0.022b |
| Pgb1-RNAi | 0.134 ± 0.028a | 0.703 ± 0.037d |
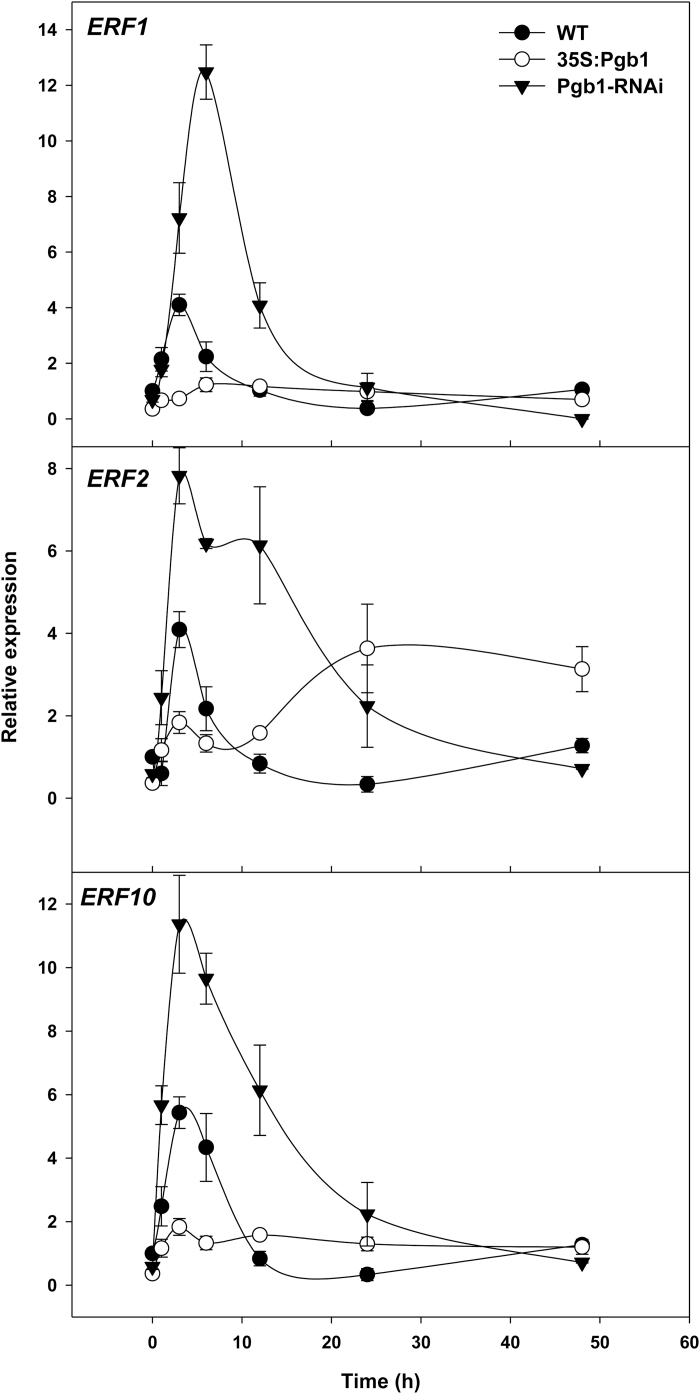
Ethylene response, estimated by the expression of ETHYLENE RESPONSIVE FACTOR1 (ERF1), ERF2, and ERF10, was also influenced by Pgbs. Relative to water-stressed WT roots, the expression of the three genes during the first hours in PEG was generally induced in roots down-regulating AtPgb1 and suppressed in roots up-regulating AtPgb1 (Fig. 4).
Fig. 4.
Expression levels of the Ethylene Responsive Factor 1 (ERF1), ERF2, and ERF10 in roots treated with 40% PEG (–1.76 MPa) of the the wild-type (WT) and lines overexpressing (35S:Pgb1) or down-regulating (Pgb1-RNAi) AtPgb1. Values are normalized to the WT at 0 h (set at 1), and are means ±SE of three biological replicates.
The requirement of ethylene for the AtPgb1 phenotype was further established pharmacologically with 1-MCP, an ethylene-perception inhibitor (Takahashi et al., 2015), partially relieving the growth retardation of the AtPgb1-suppressing roots, and ET, an ethylene-releasing agent (Zhang et al., 2010b) limiting growth of the AtPgb1 overexpressing roots (Fig. 3B). These effects were ascribed to changes in PCD patterns (see Supplementary Fig. S6).
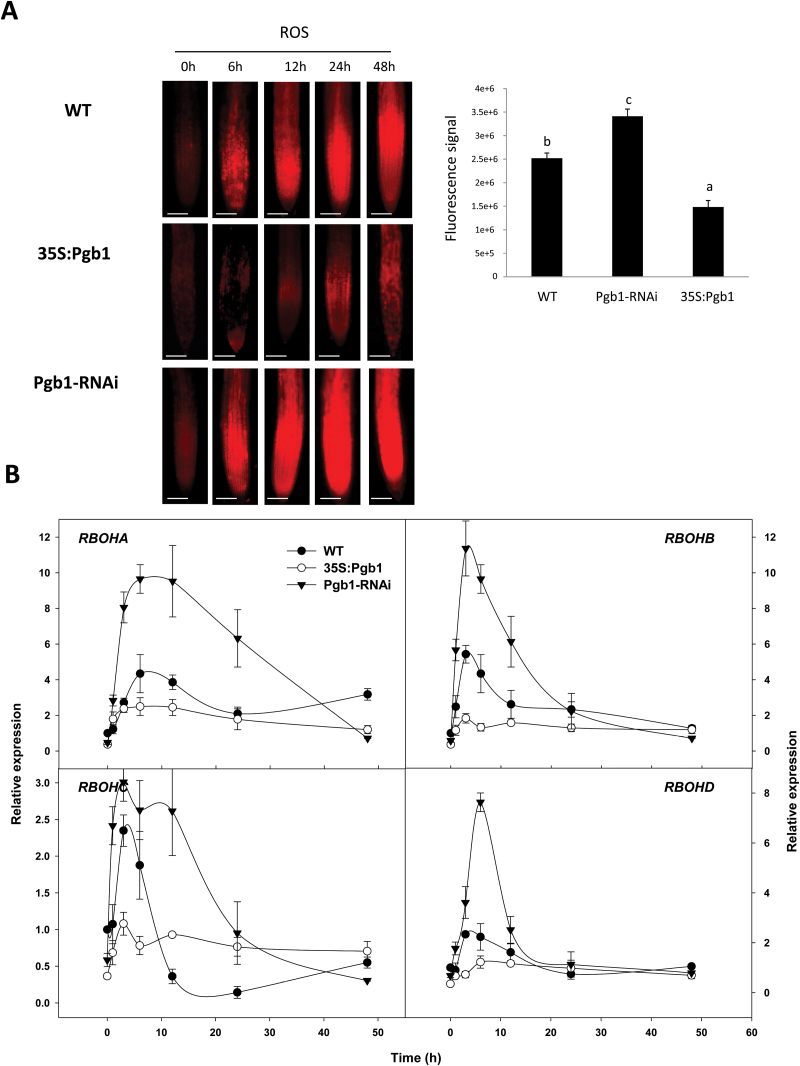
Together with ethylene, ROS act as downstream components in the AtPgb regulation of hypoxic responses (Mira et al., 2016a, 2016b). During the PEG treatment, accumulation of ROS was more pronounced in the AtPgb1-suppressing roots and less in those overexpressing AtPgb1 (Fig. 5A). Production of ROS in plant cells is mainly regulated by the activity of NADPH oxidases, a family of enzymes induced under suboptimal environmental conditions (Sagi and Fluhr, 2006). In mammals, NADPH oxidase is composed of several components, including the glycosylated transmembrane protein gp91phox (Torres and Dangl, 2005). Expression of the four Arabidopsis respiratory burst oxidases (RBOHA–D), homologs to gp91phox and reliable indicators of ROS generation (Lin et al., 2009), increased markedly in roots suppressing AtPgb1, peaking rapidly after only a few hours on PEG-medium (Fig. 5B). Relative to WT roots that exhibited a more moderate RBOH induction, roots overexpressing AtPgb1 had a generally lower expression of the same genes with the exception of RBHOC after 12 h of PEG treatment (Fig. 5B). The involvement of NADPH oxidase-generated ROS production in the cessation of root growth observed in the AtPgb1-suppressing line was demonstrated with diphenyleneiodonium (DPI), an inhibitor of NADPH-oxidase (Huang et al., 2014). Inclusion of DPI partially restored growth in water-stressed roots suppressing AtPgb1 (Fig. 3B), and this was associated with a decrease in PCD (see Supplementary Fig. S6).
Fig. 5.
Phytoglobin effects on reactive oxygen species (ROS) during water stress. (A) Localization of ROS in the wild-type (WT) and lines overexpressing (35S:Pgb1) or down-regulating (Pgb1-RNAi) AtPgb1. Roots were grown on 40% PEG (–1.76 MPa) and stained at different time points (as indicated). Scale bars are 100 μm. The graph indicates the fluorescence signal (in pixels) within the root apical tip (1mm) at 48 h. Values are means ±SE of three biological replicates. Different letters on the graph indicate statistically significant differences (P<0.05). (B) Expression levels of the Arabidopsis respiratory burst oxidases (RBOHA–D) in roots treated with 40% PEG. Values are normalized to the WT at 0 h (set at 1), and are means ±SE of three biological replicates.
In addition to demonstrating the involvement of both ethylene and ROS in the AtPgb1 regulation of root growth under PEG stress, these results, in the context of previous findings (Mira et al., 2016a, 2016c), indicate that AtPgb1 expression may act as a strategy to protect plant cells from death and to sustain growth under severe stress conditions through reduction of ROS and ethylene levels within stressed cells.
AtPgb1 is required for RAM maintenance and functionality in water-stressed roots
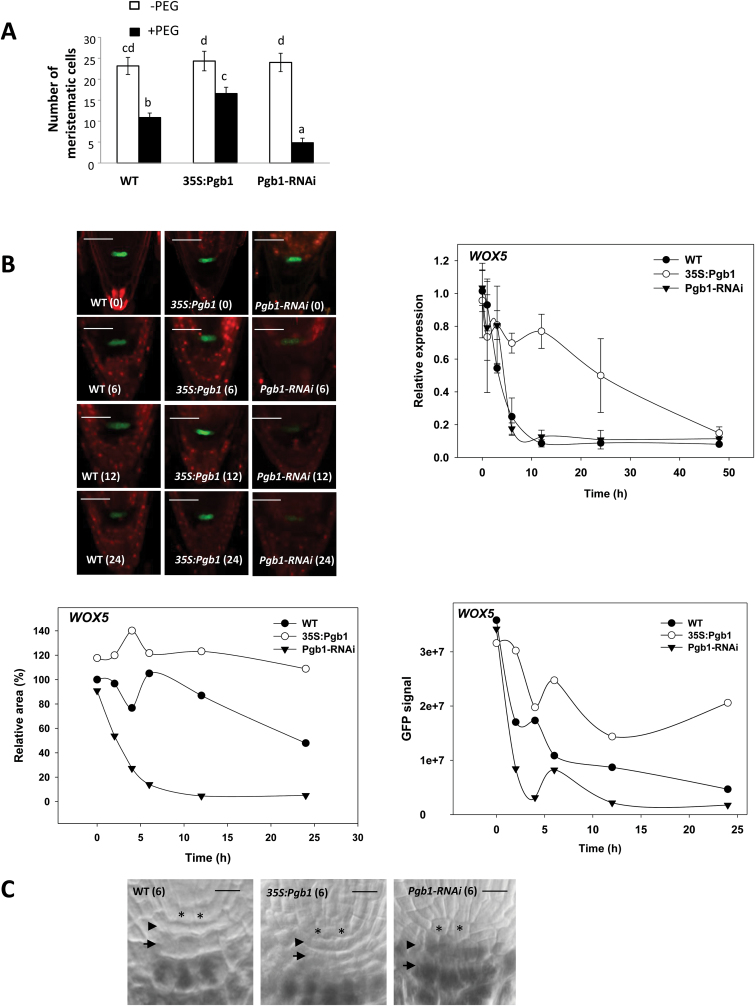
To assess if the death program in water-stressed roots is preceded by alterations in RAM patterning, the structure of the root tip in the different lines was analysed. Water stress reduced the number of root meristematic cells and this effect was attenuated in plants overexpressing AtPgb1 and was more apparent in those suppressing AtPgb1 (Fig. 6A). To establish if the reduction in meristem size was accompanied by malfunctions of the meristematic cells and to identify the exact meristematic domains influenced by AtPgb1, we crossed lines with altered levels of AtPgb1 with reporter lines (WOX5:GFP, SCR:GFP, and WER:GFP) that demark specific root domains (see Supplementary Fig. S1B). The GFP signal for WOX5, a reliable marker of the quiescent cells (QCs) in the RAM (Pi et al., 2015), declined slowly in water-stressed WT roots (Fig. 6B). In roots suppressing AtPgb1, the fluorescence almost completely disappeared after a few hours in PEG. This rapid decline in the GFP signal was accompanied by a reduction in WOX5 expression area and transcripts. PEG-treated roots overexpressing AtPgb1 retained the WOX5 signal and exhibited the highest levels of WOX5 transcripts (Fig. 6B). The function of the WOX5-expressing QCs is to act as an ‘organizing centre’ conferring ‘stem state’ to the surrounding cells, and the loss of this state, resulting from WOX5 suppression, compromises root growth (Pi et al., 2015). To assess if the rapid loss of the WOX5 signal in PEG-treated roots suppressing AtPgb1 causes premature and terminal differentiation of the distal columella stem cells, we examined the differentiation status of the columella by staining for starch granules with Lugol’s stain. This stain is routinely used to distinguish between the starchless columella stem cells (and their immediate derivatives) from the starch-containing differentiated columella cells (González-García et al., 2011). After 6 h in PEG, both WT and 35S:Pgb1 roots exhibited two starchless layers of cells below the QCs (asterisks in Fig. 6C): the columella stem cells (arrowheads) and their most immediate derivatives (arrows). This was in contrast to roots suppressing AtPgb1, which showed early signs of differentiation in the distal columella stem cells.
Fig. 6.
Analysis of meristem size and WOX5 localization and expression in water-stressed roots. (A) Number of meristematic cells in roots of the wild-type (WT) and lines overexpressing (35S:Pgb1) or down-regulating (Pgb1-RNAi) AtPgb1 grown for 48 h in the absence or presence of 40% PEG (–1.76M Pa). Values are means ±SE of three biological replicates, each consisting of at least 10 sections. Different letters indicate statistically significant differences (P<0.05). (B) Confocal images of WOX5:GFP marking the quiescent cells (QCs). Numbers in brackets on the images indicate hours of treatment in PEG. Scale bars are 50 μm. The graphs show the relative area of WOX5 expression (normalized to the WT at 0 h, which was set at 100%), the intensity of the GFP signal (in pixels), and the relative abundance of the WOX5 transcripts (normalized to the WT at 0 h, which was set at 1) in roots of the different lines grown in the presence of 40% PEG. (C) Meristems of roots cultured for 6 h in 40% PEG stained for starch granules (black precipitates) with Lugol’s solution. Quiescent cells (*), columella stem cells (arrowhead), and the most immediate columella stem cell derivatives (arrows) are shown. Scale bars are 15 μm.
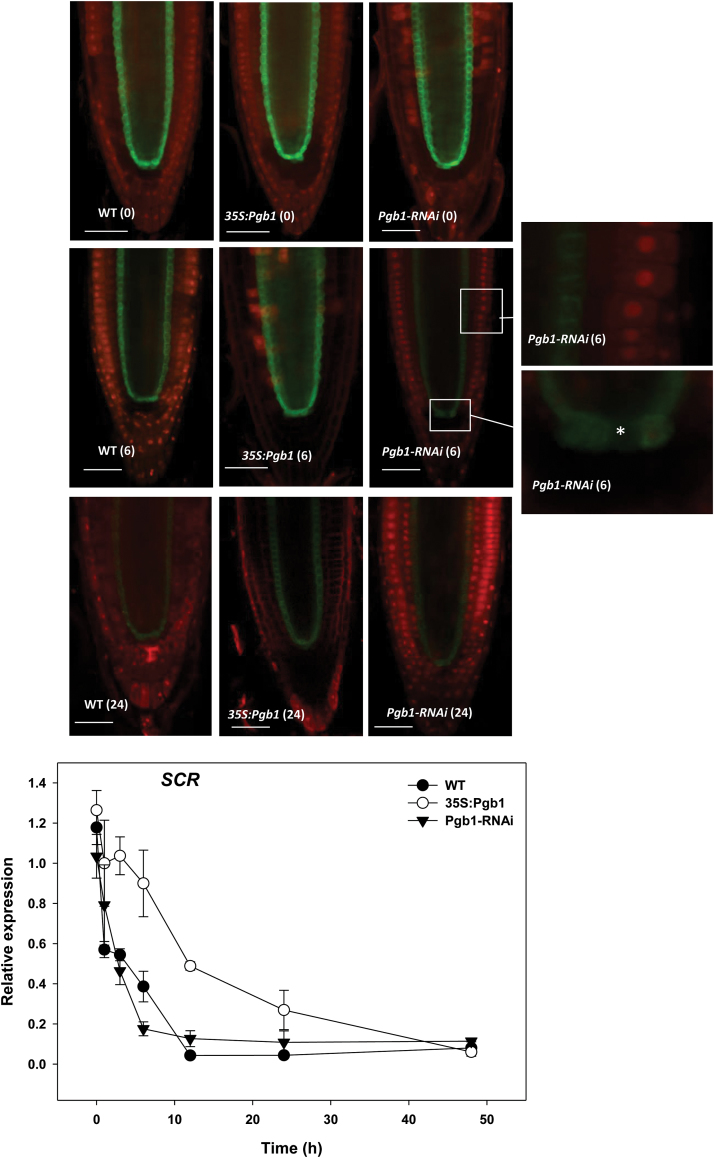
The signals of two other key root markers, SCR and WER, the first demarking the endodermis and QCs, and the second the lateral root cap (see Supplementary Fig. S1B), were also affected by water stress. The fluorescence signal and the transcript levels of SCR decreased gradually in WT roots treated with PEG (Fig. 7). This decrease was less pronounced in roots overexpressing AtPgb1, while in those suppressing AtPgb1 the signal was very weak after only 6 h in PEG (Fig. 7). QCs were the first to lose the GFP fluorescence (asterisk in Fig. 7).
Fig. 7.
Localization and expression of SCARECROW (SCR) in the wild-type (WT) and lines overexpressing (35S:Pgb1) or down-regulating (Pgb1-RNAi) AtPgb1 grown for 48 h in the absence or presence of 40% PEG (–1.76M Pa). Numbers in brackets on the images indicate hours of treatment in PEG. Confocal images of SCR:GFP marking the endodermis and quiescent cells (QCs), and the relative abundance of SCR transcripts; * indicates quiescent cell. The expression values are normalized to the WT at 0 h (set at 1), and are means ±SE of three biological replicates. Scale bars are 50 μm.
Application of PEG to WT roots resulted in a decline of WER transcripts and a concomitant loss of fluorescent signal, starting from the most mature domains of the lateral root cap (see Supplementary Fig. S8). After 24 h in PEG, the WER signal was very faint and only visible within the very innermost lateral root cap cells. A rapid decline in WER fluorescence was observed in roots suppressing AtPgb1 after only 4 h in PEG, with no signal detected in these roots after 12 h. This was in contrast to roots overexpressing AtPgb1, which exhibited the highest WER signal throughout the imposition of water stress (see Supplementary Fig. S8).
Collectively, these results demonstrate that PEG stress compromises cell fate specification and tissue patterning in the RAM, and that these effects are aggravated by suppression of AtPgb1 and alleviated in those situations where AtPgb1 is induced. Notably, suppression of AtPgb1 alters the functionality of the WOX5-expressing QCs, leading to the premature differentiation of the stem cells and a reduction in RAM size.
Auxin gradient at the root tip is influenced by AtPgb1
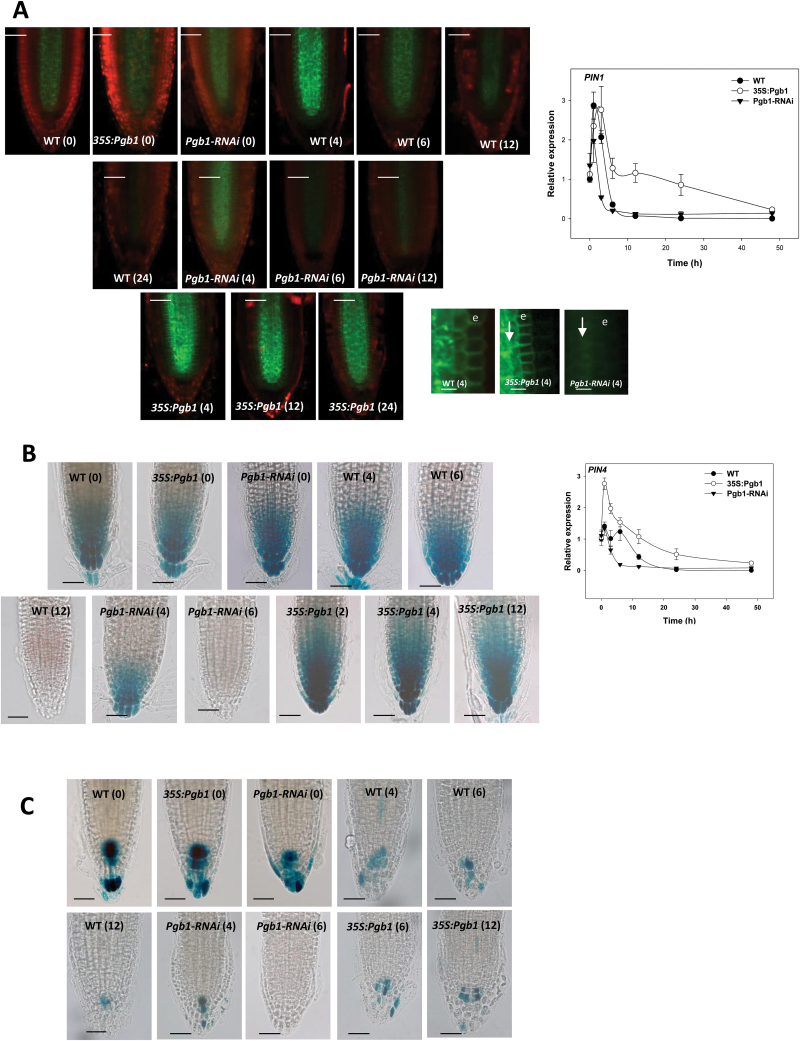
Proper functioning of the QCs is ensured by the basipetal PIN-mediated flow of auxin, which accumulates at the centre of the RAM (Vieten et al., 2005). Dissipation of auxin maxima at the root tip or interference with auxin translocation alters the behaviour of the RAM (Lee et al., 2013). Based on the premise that NO perturbs the PIN-mediated movement of auxin, producing a phenocopy of the RAM abnormalities described in the present study (Fernandez-Marcos et al., 2011), and that AtPgb1 is an effective NO scavenger (Hebelstrup et al., 2006), auxin flow and accumulation were monitored in PEG-treated roots with altered levels of AtPgb1. The pattern of auxin flow was estimated by analysing the expression and localization of PIN1 (using a PIN1-GFP construct), the auxin efflux factor responsible for the basipetal translocation of auxin in the stele and endodermis (Vieten et al., 2005), and PIN4 (using the PIN4:GUS construct), which regulates auxin movement and redistribution at the root tip (Friml et al., 2002) (see Supplementary Fig. S1A). Auxin maxima were visualized by the activity of DR5:GUS (Ni et al., 2001).
The expression of PIN1 was induced in all lines during the first hours in PEG and then it declined, especially in WT and AtPgb1-suppressing roots (Fig. 8A). Roots overexpressing AtPgb1 retained the highest levels of PIN1 transcripts. This expression pattern was confirmed by the localization signal which, relative to the WT, was very strong in the roots overexpressing AtPgb1 throughout the course of the experiment. A sharp decline in PIN1-GFP fluorescence was observed in AtPgb1-suppressing roots after only 4 h in PEG, especially within the endodermis, the first tissue to lose the signal (‘e’ in Fig. 8A).
Fig. 8.
Auxin flow and accumulation during PEG-induced water stress in the wild-type (WT) and lines overexpressing (35S:Pgb1) or down-regulating (Pgb1-RNAi) AtPgb1 grown for 48 h in the absence or presence of 40% PEG (–1.76M Pa). Numbers in brackets on the images indicate hours of treatment in PEG. (A) Confocal images of PIN1-GFP, and relative abundance of PIN1 transcripts during the imposition of water stress. Arrows indicate the basipetal flow of auxin; e, endodermis. Scale bars are 50 μm (larger panels) and 5 μm (smaller panels). (B) Expression of PIN4:GUS and relative abundance of PIN4 transcripts in roots. The relative expression values in (A) and (B) are normalized to the WT at 0 h (set at 1), and are means ±SE of three biological replicates. Scale bars in (B) are 50 μm. (C) Localization of DR5:GUS in roots of the different lines subjected to PEG-induced water stress. Scale bars are 50 μm.
After an initial induction, most pronounced in the roots overexpressing AtPgb1, the levels of PIN4 transcripts decreased during the imposition of PEG stress (Fig. 8B). During the first 24 h this decrease was inversely related to the amount of AtPgb1 present in the tissue. Localization studies confirmed this trend and showed that differences among lines occurred after 4 h in PEG when the AtPgb1-suppressing roots displayed a fainter GUS signal, which was completely lost after 6 h in PEG. This was in contrast to roots up-regulating AtPgb1 in which a strong signal was retained even at 12 h in PEG, when no GUS was detected in the WT roots (Fig. 8B).
The auxin signal, visualized by the activity of DR5:GUS, was strong in WT roots at the beginning of the experiment (0 h in PEG), but dissipated gradually during water stress (Fig. 8C). After 12 h only a few cells in the centre of the RAM accumulated auxin. Relative to the WT, the GUS domain in the AtPgb1-suppressing roots was reduced after only 4 h in PEG, and was completely lost during the following two hours. A significant number of GUS-stained cells were still observed in the centre of the RAM of roots up-regulating AtPgb1, even after several hours of water stress (Fig. 8C).
Taken together, these findings suggest that PEG stress alters the PIN-mediated accumulation of auxin at the root tip, and they demonstrate that the presence of AtPgb1 is required to attenuate these changes. A rapid decline in PIN1 and PIN4 occurs in AtPgb1 down-regulating roots after only a few hours of water deficit and this is accompanied by the dissipation of the auxin maxima within the RAM.
Discussion
Short or extended periods of severe water deficit can compromise plant growth and induce death of root cells (Ramachandra Reddy et al., 2004; Duan et al., 2010). Through the use of PEG, it has been previously demonstrated that the deleterious effects of drought on root growth are the result of premature differentiation of the RAM (Ji et al., 2014), and extensive death at the root tip (Duan et al., 2010). The recurrence of these events during several types of stress reveals the susceptibility of roots to adverse environmental conditions. It has been suggested that Pgbs might exercise a protective role that allows roots to cope with diverse types of stress (Mira et al., 2016b). For example, the hypoxia-inhibition of root growth resulting from differentiation and death of root cells was attenuated by the overexpression of Pgbs and aggravated when Pgbs were down-regulated (Mira et al., 2016b). Based on these observations, the present work examined the involvement of Pgbs on death processes in root cells triggered by lethal applications of PEG.
The Arabidopsis AtPgb1, rapidly induced in roots at the onset of water stress (Fig. 1B), influenced root responses to water deficit, with high levels (35S:Pgb1 line) alleviating the PEG-growth inhibition, and low levels (Pgb1-RNAi) accentuating the inhibition (Fig. 1A). The protective role of AtPgb1 during water deficit appears to be linked to the ability of the protein to reduce PCD at the root tip (Fig. 2), possibly by limiting ethylene synthesis (Table 1), ethylene response (estimated by the expression levels of ERF1, 2, and 10) (Fig. 4), and accumulation of ROS (Fig. 5), all of which are known effectors of the death program. The involvement of both ethylene and ROS in the AtPgb1 response was demonstrated pharmacologically, with 1-MCP, an inhibitor of ethylene perception (Takahashi et al., 2015), and DPI, an inhibitor of NADPH-oxidase (Bindschedler et al., 2006) partially rescuing growth in AtPgb1-suppressing roots, and ET, an ethylene-releasing agent (Zhang et al., 2010b) suppressing growth in roots over-expressing AtPgb1 (Fig. 3B). These phenotypes were associated with changes in PCD patterns as a result of the pharmacological treatments (see Supplementary Fig. S6).
Programmed cell death in plant cells can be triggered by many signals, some of which originate from the endoplasmic reticulum (ER) (Zuppini et al., 2004; Watanabe and Lam, 2008). One of the key functions of the ER is to regulate protein maturation and maintain a proper equilibrium between unfolded and folded proteins. Alteration of this equilibrium, elevating the levels of unfolded or misfolded proteins, triggers transduction pathways culminating with apoptosis in animals and PCD in plants (Boyce and Yuan, 2006; Watanabe and Lam, 2008). Evidence from our current study supports the findings of Duan et al. (2010) that documented ER-induced PCD in water-stressed tissue, and further suggests that AtPgb1 might reduce the death program by alleviating ER stress through the activation of the unfolded protein response (UPR). This UPR is highly conserved among species (Boyce and Yuan, 2006) and involves the participation of AtBI-1, AtBiP2, and AtPR1, attenuators of ER stress that were all induced in roots overexpressing AtPgb1 and suppressed in roots down-regulating AtPgb1 (Fig. 3A). Often referred to as ‘survival factor’, the ER-located AtBI-1 suppresses the BAX (pro-apoptotic protein) activation of cell death in mammalian cells (Huckelhoven, 2004). Overexpression of AtBI-1 in plants alleviates ER stress and reduces PCD progression, while its suppression accelerates the death program (Watanabe and Lam, 2006, 2008). The AtPgb1 regulation of AtBiP2 and AtPR1 was consistent with that of AtBI-1. AtBiP2 is a HSP70-type of chaperon involved in the secretion of proteins in the ER and a marker of UPR induction in eukaryotes (Koizumi et al., 2001), while AtPR1 is a pathogenesis-related protein and a downstream intermediate of ER signalling (Watanabe and Lam, 2008). The contribution of ER-stress to the death program was further confirmed by the ability of PBA, a chaperon reducing ER stress through the stabilization of protein conformation (Watanabe and Lam, 2008), to alleviate the PEG-induced retardation of growth (Fig. 3B) and to suppress PCD (see Supplementary Fig. S6).
Commencement of the death program is often preceded by cellular differentiation (Ramsdale, 2012), a premature event observed in water-stressed wheat roots (Ji et al., 2014), that can alter cell fate and tissue patterning in the RAM (Shishkova et al., 2008). In Arabidopsis, the RAM surrounds the QCs, comprising mitotically inactive cells with the unique function to maintain the proximally and laterally situated stem cells in an undifferentiated state (Dolan et al., 1993 and Supplementary Fig. S1A). The delicate balance between the rate of proliferation of the stem cells and the differentiation of their derivatives, ensuring a constant meristem size, makes the RAM extremely vulnerable to environmental perturbations. Evidence presented in this paper suggests that during water deficit AtPgb1 expression contributes to the persistence of a functional meristem through the specification of the WOX5-expressing QCs. The retention of WOX5 expression observed in the AtPgb1-overexpressing roots subjected to PEG stress (Fig. 6B) ensures the proper function of the QCs as the ‘organizing centre’ of the RAM, thus preserving the size (Fig. 6A) and function of the RAM. In contrast, suppression of AtPgb1 resulted in the premature loss of WOX5 expression after only a few hours in PEG, causing the differentiation of columella stem cells that accumulate starch granules (Fig. 6C). The loss of pluripotency and differentiation of the stem cells is a plausible cause of the smaller RAM observed in the AtPgb1-suppressing roots (Fig. 6A). Through the specification of QC identity, WOX5 acts as a regulator of stem cell maintenance by repressing the differentiation factor CDF4 in the adjacent stem cells (Pi et al., 2015). Consistent with our results, the ablation of the QCs (van den Berg et al., 1997) or the loss of function associated with the wox5 mutant (Sarkar et al., 2007) cause the differentiation of the subtending columella initials, while ectopic WOX5 expression is sufficient to reprogram fully differentiated columella cells into stem cells (Pi et al., 2015). The rapid, PEG-induced loss of root tissue specification, aggravated by suppression of AtPgb1 and delayed by its up-regulation, was also apparent through the localization of SCR (Fig. 7). In addition to participating in radial root patterning by controlling the asymmetric cell division pattern of the daughter of the cortex–endodermis initial (Di Laurenzio et al., 1996), SCR is also required for proper QC activity (Sabatini et al., 2003). The early (6 h in PEG) suppression of SCR in the AtPgb1-suppressing roots, especially in the QCs (asterisk in Fig. 7), reinforces the argument for the requirement of AtPgb1 in delaying the loss of QC specification during water deficit. Together with cell specification and tissue patterning in the central domains of the RAM, it cannot be excluded that AtPgb1 delays the loss of tissue identity in the periphery of the root tip, as evidenced by the changes in expression and localization patterns of the lateral root cap marker WER (see Supplementary Fig. S8). The pattern of cellular proliferation, estimated by the GUS activity of two cyclins (cycB3,1:GUS in the meristematic region and cycA1,3:GUS in mature tissue; see Supplementary Fig. S9), further confirms the requirement of AtPgb1 for normal root growth under conditions of water deficit.
Maintenance and function of the RAM is controlled by the PIN-mediated accumulation of auxin at the root tip, and alterations in auxin flow by high levels of NO lead to the same meristem abnormalities observed in our study, namely a decrease in meristem size and premature differentiation of meristematic cells (Fernandez-Marcos et al., 2011). This observation, in conjunction with the well-documented ability of AtPgb1 to scavenge NO (Hebelstrup et al., 2006), prompted us to determine whether the early meristematic defects observed in water-stressed AtPgb1-suppressing roots were associated with perturbations in the flow and accumulation of auxin in the RAM. The patterns of PIN1 and 4 expression and localization were rapidly altered in tissue suppressing AtPgb1 after only a few hours in PEG (Fig. 8A, B). These alterations, denoting changes in the long-range basipetal canalization of auxin mediated by PIN1 (Petersson et al., 2009) and its maintenance and redistribution within the meristematic cells, regulated by PIN4 (Friml et al., 2002), reflect the rapid (6 h in PEG) disappearance of auxin in the RAM (Fig. 8C). Abnormal root tips have been associated with defective PIN1 localization, while misexpression of PIN4 causes deviations in QC fate (Friml et al., 2003). The retention of a PIN-directed auxin maxima at the RAM, and specifically in the QCs (Petersson et al., 2009), is paramount for meristem functionality and, most importantly, for modulating cell fate regulators, including WOX5 (Ding and Friml, 2010) and SCR (Sabatini et al., 1999). Mutants exhibiting decreasing DR5:GUS activity displayed aberrations in cell specification and tissue patterning (Sabatini et al., 1999).
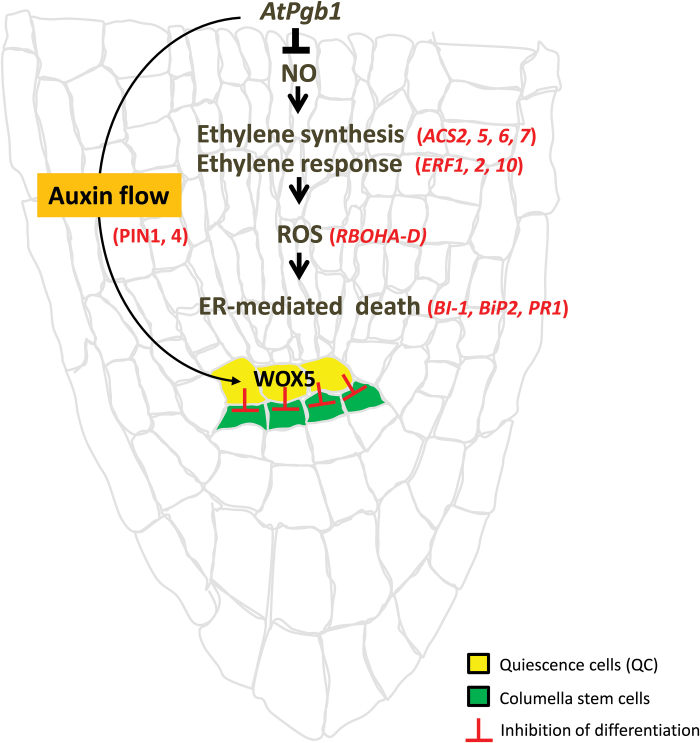
In conclusion, while not producing any visible root phenotype in the absence of PEG, the expression level of AtPgb1 influences the root response to severe PEG stress. By modulating ethylene and ROS, AtPgb1 alleviates ER stress-induced death in PEG-stressed roots and delays the degradation of the RAMs by maintaining meristem functionality and fate identity in the WOX-5-expressing QCs. These events are most likely associated with the retention of the flow of auxin and its accumulation at the root tip (Fig. 9). The similarity of regulatory components shared by the root and shoot apical meristems, where the WOX-5 like WUSCHEL gene possesses stem cell-promoting functions in the shoot, as well as the presence of AtPgb1 in the shoot tip, suggest a potential role of this protein in protecting meristematic cells during conditions of extreme stress.
Fig. 9.
Proposed model of AtPgb1 action during severe water stress. The model is based on the current work and integrates some information from previous studies (Mira et al., 2016b). By suppressing production of nitric oxide (NO), AtPgb1 reduces the promotive effect of ethylene and ROS on ER-mediated death of root cells. AtPgb1 is also required to maintain the fate of the QCs, which prevent the differentiation of the subtending columella stem cells. This effect, possibly mediated by the flow and distribution of auxin, contributes to the retention of a functional RAM during conditions of PEG stress. Genes whose activities have been measured are listed in red.
Supplementary Data
Supplementary data are available at JXB online.
Fig. 1. Tissue patterning in the Arabidopsis root apical meristem.
Fig. 2. Water potentials of agar media infiltrated with PEG.
Fig. 3. Water stress inhibition of root growth.
Fig. 4. Time course of root elongation on different media.
Fig. 5. Expression of AtPgb1 in wild-type roots grown with different water potentials.
Fig. 6. PCD in root tips of PEG-stressed roots.
Fig. 7. Alteration of expression of ethylene-biosynthetic genes by phytoglobin.
Fig. 8. WER expression and localization.
Fig. 9. Cell division patterns in PEG-treated roots.
Table S1. List of primers used in this study.
Supplementary Material
Acknowledgements
This work was supported by a NSERC Discovery Grant to CS. The authors thank the following colleagues for providing the Arabidopsis lines: Dr K. Hebelstrup (35S:Pgb1 and Pgb1-RNAi), Dr P. Benfey (SCR:GFP, and WER:GFP), and Dr O. Lorenzo (PIN1-GFP); and AgroFresh Inc. for providing 1-MCP. The valuable technical assistance of Mr D. Durnin is also acknowledged.
References
- Bengough AG, McKenzie BM, Hallett PD, Valentine TA. 2011. Root elongation, water stress, and mechanical impedance: a review of limiting stresses and beneficial root tip traits. Journal of Experimental Botany 62, 59–68. [DOI] [PubMed] [Google Scholar]
- Bindschedler LV, Dewdney J, Blee KA et al. 2006. Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. The Plant Journal 47, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert HJ, Sheveleva E. 1998. Plant stress adaptations–making metabolism move. Current Opinion in Plant Biology 1, 267–274. [DOI] [PubMed] [Google Scholar]
- Boyce M, Yuan J. 2006. Cellular response to endoplasmic reticulum stress: a matter of life or death. Cell Death and Differentiation 13, 363–373. [DOI] [PubMed] [Google Scholar]
- Bulankova P, Akimcheva S, Fellner N, Riha K. 2013. Identification of Arabidopsis meiotic cyclins reveals functional diversification among plant cyclin genes. PLoS Genetics 9, e1003508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves MM, Pereira JS, Maroco J, Rodrigues ML, Ricardo CP, Osório ML, Carvalho I, Faria T, Pinheiro C. 2002. How plants cope with water stress in the field. Photosynthesis and growth. Annals of Botany 89, 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. 1996. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433. [DOI] [PubMed] [Google Scholar]
- Ding Z, Friml J. 2010. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proceedings of the National Academy of Science, USA 107, 12046–12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84. [DOI] [PubMed] [Google Scholar]
- Dordas C, Hasinoff BB, Igamberdiev AU, Manac’h N, Rivoal J, Hill RD. 2003. Expression of a stress-induced hemoglobin affects NO levels produced by alfalfa root cultures under hypoxic stress. The Plant Journal 35, 763–770. [DOI] [PubMed] [Google Scholar]
- Duan Y, Zhang W, Li B, Wang Y, Li K, Sodmergen, Han C, Zhang Y, Li X. 2010. An endoplasmic reticulum response pathway mediates programmed cell death of root tip induced by water stress in Arabidopsis. New Phytologist 186, 681–695. [DOI] [PubMed] [Google Scholar]
- Fernandez-Marcos M, Sanz L, Lewis DR, Muday GK, Lorenzo O. 2011. Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED 1 (PIN1)-dependent acropetal auxin transport. Proceedings of the National Academy of Science, USA 108, 18506–18511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Benková E, Blilou I et al. 2002. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108, 661–673. [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. 2003. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 426, 147–153. [DOI] [PubMed] [Google Scholar]
- González-García MP, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-García S, Russinova E, Caño-Delgado AI. 2011. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138, 849–859. [DOI] [PubMed] [Google Scholar]
- Gunawardena AH, Greenwood JS, Dengler NG. 2004. Programmed cell death remodels lace plant leaf shape during development. The Plant Cell 16, 60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebelstrup KH, Hunt P, Dennis ES, Jensen SB, Jensen EØ. 2006. Hemoglobin is essential for normal growth of Arabidopsis organs. Physiologia Plantarum 127, 157–166. [Google Scholar]
- Hill R, Hargrove M, Arredondo-Peter R. 2016. Phytoglobin: a novel nomenclature for plant globins accepted by the globin community at the 2014 XVIII conference on Oxygen-Binding and Sensing Proteins. F1000Research 5, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RD. 2012. Non-symbiotic haemoglobins—What’s happening beyond nitric oxide scavenging?AoB Plants 2012, pls004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JH, Chu H, Zhang C, Ghosh D, Gong X, Xu J. 2015. A quantitative analysis of stem cell homeostasis in the Arabidopsis columella root cap. Frontiers in Plant Science 6, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Hill RD, Wally OS, Dionisio G, Ayele BT, Jami SK, Stasolla C. 2014. Hemoglobin control of cell survival/death decision regulates in vitro plant embryogenesis. Plant Physiology 165, 810–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckelhoven R. 2004. BAX Inhibitor-1, an ancient cell death suppressor in animals and plants with prokaryotic relatives. Apoptosis 9, 299–307. [DOI] [PubMed] [Google Scholar]
- Hunt PW, Klok EJ, Trevaskis B, Watts RA, Ellis MH, Peacock WJ, Dennis ES. 2002. Increased level of hemoglobin 1 enhances survival of hypoxic stress and promotes early growth in Arabidopsis thaliana. Proceedings of the National Academy of Science, USA 99, 17197–17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Liu L, Li K, Xie Q, Wang Z, Zhao X, Li X. 2014. PEG-mediated osmotic stress induces premature differentiation of the root apical meristem and outgrowth of lateral roots in wheat. Journal of Experimental Botany 65, 4863–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N, Martinez IM, Kimata Y, Kohno K, Sano H, Chrispeels MJ. 2001. Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiology 127, 949–962. [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Lee WS, Kim SH. 2013. Hormonal regulation of stem cell maintenance in roots. Journal of Experimental Botany 64, 1153–1165. [DOI] [PubMed] [Google Scholar]
- Leite M, Quinta-Costa M, Leite PS, Guimaraes JE. 1999. Critical evaluation of techniques to detect and measure cell death – study in a model of UV radiation of the leukaemic cell line HL60. Analytical Cellular Pathology 19, 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Zhu C, Gan L, Ng D, Xia K. 2015. GA3 enhances root responsiveness to exogenous IAA by modulating auxin transport and signalling in Arabidopsis. Plant Cell Reports 34, 483–494. [DOI] [PubMed] [Google Scholar]
- Lin F, Ding H, Wang J, Zhang H, Zhang A, Zhang Y, Tan M, Dong W, Jiang M. 2009. Positive feedback regulation of maize NADPH oxidase by mitogen-activated protein kinase cascade in abscisic acid signalling. Journal of Experimental Botany 60, 3221–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mira M, Hill RD, Stasolla C. 2016a. Regulation of programmed cell death by phytoglobins. Journal of Experimental Botany 67, 5901–5908. [DOI] [PubMed] [Google Scholar]
- Mira MM, El-Khateeb EA, SayedAhmed HI, Hill RD, Stasolla C. 2017. Are avoidance and acclimation responses during hypoxic stress modulated by distinct cell-specific mechanisms?Plant Signaling & Behavior 12, e1273304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira MM, Hill RD, Stasolla C. 2016b. Phytoglobins improve hypoxic root growth by alleviating apical meristem cell death. Plant Physiology 172, 2044–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira MM, Wally OS, Elhiti M, El-Shanshory A, Reddy DS, Hill RD, Stasolla C. 2016c. Jasmonic acid is a downstream component in the modulation of somatic embryogenesis by Arabidopsis Class 2 phytoglobin. Journal of Experimental Botany 67, 2231–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni DA, Wang LJ, Ding CH, Xu ZH. 2001. Auxin distribution and transport during embryogenesis and seed germination of Arabidopsis. Cell Research 11, 273–278. [DOI] [PubMed] [Google Scholar]
- Ning SB, Wang L, Song YC. 2002. Identification of programmed cell death in situ in individual plant cells in vivo using a chromosome preparation technique. Journal of Experimental Botany 53, 651–658. [DOI] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M et al. 2009. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. The Plant Cell 21, 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN. 2012. Control of Arabidopsis root development. Annual Review of Plant Biology 63, 563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, Groot E, Laux T. 2015. Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Developmental Cell 33, 576–588. [DOI] [PubMed] [Google Scholar]
- Ramachandra Reddy A, Chaitanya KV, Vivekanandan M. 2004. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. Journal of Plant Physiology 161, 1189–1202. [DOI] [PubMed] [Google Scholar]
- Ramsdale M. 2012. Programmed cell death in the cellular differentiation of microbial eukaryotes. Current Opinion in Microbiology 15, 646–652. [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Ercoli MF, Debernardi JM et al. 2015. MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. The Plant Cell 27, 3354–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H et al. 1999. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. 2003. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes & Development 17, 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi M, Fluhr R. 2006. Production of reactive oxygen species by plant NADPH oxidases. Plant Physiology 141, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. 2007. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814. [DOI] [PubMed] [Google Scholar]
- Shishkova S, Rost TL, Dubrovsky JG. 2008. Determinate root growth and meristem maintenance in angiosperms. Annals of Botany 101, 319–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Sachs MM. 2003. Molecular and cellular adaptations of maize to flooding stress. Annals of Botany 91, 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Yamauchi T, Rajhi I, Nishizawa NK, Nakazono M. 2015. Transcript profiles in cortical cells of maize primary root during ethylene-induced lysigenous aerenchyma formation under aerobic conditions. Annals of Botany 115, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Dangl JL. 2005. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Current Opinion in Plant Biology 8, 397–403. [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. 1997. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390, 287–289. [DOI] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benkova E, Benjamins R, Beeckman T, Luschnig C, Friml J. 2005. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132, 4521–4531. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Lam E. 2006. Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. The Plant Journal 45, 884–894. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Lam E. 2008. BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. The Journal of Biological Chemistry 283, 3200–3210. [DOI] [PubMed] [Google Scholar]
- Welch WJ, Brown CR. 1996. Influence of molecular and chemical chaperones on protein folding. Cell Stress & Chaperones 1, 109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westgate ME, Boyer JS. 1985. Osmotic adjustment and the inhibition of leaf, root, stem and silk growth at low water potentials in maize. Planta 164, 540–549. [DOI] [PubMed] [Google Scholar]
- Williams B, Verchot J, Dickman MB. 2014. When supply does not meet demand-ER stress and plant programmed cell death. Frontiers in Plant Science 5, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Han W, De S et al. 2010a. ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. The Plant Journal 64, 764–774. [DOI] [PubMed] [Google Scholar]
- Zhang W, Hu W, Wen CK. 2010b. Ethylene preparation and its application to physiological experiments. Plant Signaling & Behavior 5, 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jiao Y, Liu Z, Zhu YX. 2015. ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells. Nature Communications 6, 6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Gu RL, Gao P, Wang GY. 2008. A nonsymbiotic hemoglobin gene from maize, ZmHb, is involved in response to submergence, high-salt and osmotic stresses. Plant Cell Tissue and Organ Culture 95, 227–237. [Google Scholar]
- Zuppini A, Navazio L, Mariani P. 2004. Endoplasmic reticulum stress-induced programmed cell death in soybean cells. Journal of Cell Science 117, 2591–2598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.