We investigated the impact of meningococcal conjugate vaccine on carriage of Neisseria meningitidis in university students. Expansion of capsule-expressing isolates from the 2013-strain of serogroup W clonal complex sequence type 11 but not isolates from serogroup Y clonal complex 23 suggests differential susceptibilities to vaccine-induced immunity.
Keywords: Neisseria, meningitidis, whole, genome sequencing, carriage, serogroup W, serogroup Y, epidemiology, vaccine
Abstract
Background
In the United Kingdom, rising levels of disease due to Neisseria meningitidis serogroup W clonal complex (cc) sequence type (ST) 11 (MenW:cc11) strains led to introduction of meningococcal conjugate vaccine (MenACWY) for teenagers. We investigated the impact of immunization on carriage of meningococci targeted by the vaccine, using whole-genome sequencing of isolates recovered from a cohort of vaccinated university students.
Methods
Strain designation data were extracted from whole-genome sequencing data. Genomes from carried and invasive MenW:cc11 strains were compared using a gene-by-gene approach. Serogrouping identified isolates expressing capsule antigens targeted by the vaccine.
Results
Isolates with a W: P1.5,2: F1-1: ST-11 (cc11) designation and belonging to the emerging 2013-strain of the South American–United Kingdom MenW:cc11 sublineage were responsible for an increase in carried group W strains. A multifocal expansion was evident, with close transmission networks extending beyond individual dormitories. Carried group Y isolates were predominantly from cc23 but showed significant heterogeneity, and individual strain designations were only sporadically recovered. No shifts toward acapsulate phenotypes were detected in targeted meningococcal populations.
Conclusions
In a setting with high levels of MenACWY use, expansion of capsule-expressing isolates from the 2013-strain of MenW:cc11 but not MenY:cc23 isolates is indicative of differential susceptibilities to vaccine-induced immunity.
Neisseria meningitidis is a commensal of the human oropharynx that can cause invasive meningococcal disease (IMD), principally characterized by sepsis and/or meningitis [1]. The majority of IMD worldwide is caused by isolates expressing the polysaccharide capsules that define serogroups A, B, C, W, Y, and X, whereas carriage isolates are frequently acapsulate because of the inactivation or absence of genes involved in capsule expression [2–4]. The incidence and contribution of different meningococcal lineages to the overall burden of IMD varies geographically, temporally, and by age group and can be further influenced by vaccination; capsule polysaccharide–based vaccines are available against serogroups A, C, W, and Y, whereas protein-based vaccines are available against serogroup B isolates [5]. Monitoring trends in meningococcal populations requires discriminatory typing strategies. Whole-genome sequence (WGS) analyses are now routine and enable differentiation of clonal complex (CC), sequence type (ST), and even highly similar clones, thereby facilitating detection of population level trends and individual transmission events [6].
Over the last 2 decades, multiple countries have experienced increases in the incidence of IMD due to serogroup W (MenW) meningococci belonging to the ST-11 cc (cc11) [7–11]. Analysis of WGS data has indicated that most MenW:cc11 isolates belong to cc11 lineage 11.1, with the global increases in MenW:cc11 disease resulting from emergence of 2 diversifying sublineages [12]. The Hajj sublineage comprises 3 main clusters of isolates (strains), corresponding to those of the Hajj outbreak of 2000 onward (Anglo-French Hajj strain), expansion of endemic disease in South Africa from 2003 (endemic South African strain), and epidemics in sub-Saharan Africa (Burkina Faso/North African strain) [12]. The South American–United Kingdom sublineage comprises MenW:cc11 that emerged in South America and subsequently spread to the United Kingdom and Europe [12]. This second lineage is associated with atypical clinical presentation, including gastrointestinal symptoms, and a high case-fatality rate [13, 14]. Ongoing genomic surveillance has revealed further population structure details for the South American–United Kingdom MenW:cc11 sublineage, such as the initial emergence of the original United Kingdom strain in 2009, followed by subsequent emergence of the novel 2013-strain from this original United Kingdom strain [15].
The year-on-year increase in MenW:cc11 IMD cases in the United Kingdom since 2009 led to an emergency immunization program, with meningococcal ACWY conjugate vaccine (MenACWY) being recommended for adolescents. This program included a phased catch-up campaign for individuals 14–18 years of age and began in August 2015 [16]. Older adolescents and young adults were targeted because these age groups exhibit higher oropharyngeal carriage rates than other age groups, owing to social factors [17, 18]. Particularly high carriage rates are evident in young adult populations residing in semiclosed communities (eg, university students), where the potential for person-to-person transmission is especially high and can lead to isolated clusters or outbreaks of meningococcal disease [19–22]. Hence, MenACWY was also offered to new university entrants <25 years of age. Furthermore, from previous experience with meningococcal group C conjugate vaccines, targeting adolescents and young adults could result in sustained decreases in disease incidence in all age groups (ie, herd protection) by reducing the acquisition of meningococcal carriage [23].
At the University of Nottingham (Nottingham, United Kingdom), a campus-based vaccination campaign targeting freshmen in September 2015 increased MenACWY coverage in this specific student population from 31% to 71% [24]. To determine the effect of this vaccination campaign on meningococcal carriage, we conducted a cross-sectional study at the University of Nottingham, from September 2015 through March 2016 [25]. The overall meningococcal carriage rate increased throughout the study, in line with previous university-based carriage studies [26–28]. No significant change in carriage of MenY organisms occurred, but we detected a rapid and significant rise in carriage of MenW strains with PorB serotypes and porA and fHbp STs that matched alleles harbored by endemic United Kingdom MenW:cc11 invasive isolates [25]. Here we analyze whole-genome data to define the specific MenW, MenY, and nongroupable lineages present in this student cohort; investigate the genetic relatedness of carried MenW:cc11 to contemporary invasive isolates; and consider the potential mechanisms by which vaccine-targeted isolates may escape immune responses elicited by vaccination with the MenACWY capsule-based conjugate vaccine.
METHODS
Carriage Isolates
A total of 174 meningococcal isolates, all obtained from oropharyngeal carriers in 2015–16 at the University of Nottingham (East Midlands) [25], were included in the WGS analysis (Supplementary Table 1). Of these, 49 were MenW and 32 were MenY, together accounting for approximately 95% of MenW and MenY isolated during the carriage study [25]. A further 93 isolates were nongroupable (ie, they lacked ctrA or carried the capsule null locus), corresponding to approximately 70% of nongroupable isolates obtained in the 2015–2016 University of Nottingham study [25]. All isolates were chosen as known MenW, MenY, or nongroupable organisms, based on polymerase chain reaction (PCR) typing methods, without prior knowledge of their clonal complex. The Meningococcal Reference Unit, Public Health England (Manchester, United Kingdom), performed serogrouping of MenY carriage isolates, using a dot-blot enzyme-linked immunosorbent assay. Serogrouping of MenW carriage isolates was reported previously [25]. χ2 tests for significance were performed by using STATCALC (Epi Info, version 7.2.0.1; Centers for Disease Control and Prevention, Atlanta, GA).
Genomic DNA Extraction, Sequencing, Assembly, and Deposition
Meningococci were grown overnight on Columbia agar with chocolate horse blood (Thermo Fisher Scientific) at 37°C in an atmosphere of air plus 5% CO2. Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega) according to the manufacturer’s instructions. Index-tagged Illumina sequencing libraries were generated according to the manufacturer’s instructions, with an average insert size of 420 base pairs (bp). These were multiplexed and sequenced on Illumina HiSeq 2500 machines to generate 125-bp paired-end sequences. An average of 2773968 reads per sample was generated, giving an average 125-fold coverage of the N. meningitidis genome. Short-read sequences were trimmed with Trimmomatic v0.32 [29] and assembled with SPAdes v3.9.0 [30], using the recommended parameters. Assemblies were deposited in and subsequently automatically annotated by the PubMLST.org/neisseria database, which implements the Bacterial Isolate Genome Sequence (BIGSdb) platform [31]. Short-read sequences were also deposited in the European Nucleotide Archive (Supplementary Table 1).
Genomic Analyses
Isolate capsular groups, multilocus STs, and porin A and ferric enterochelin receptor types were identified from whole-genome data. For MenW:cc11 carriage isolates, population-wide genomic analyses were undertaken using the BIGSdb Genome Comparator tool implemented within the PubMLST.org/neisseria database, using the N. meningitidis cgMLST v1.0 core genome scheme (1605 loci) and default settings [31]. Output distance matrices (Nexus format) were used to generate NeighborNet networks, using SplitsTree4 (v4.14.5). WGS data from MenW:cc11 carriage isolates were analyzed in conjunction with 2 other WGS data sets: (1) all United Kingdom MenW:cc11 invasive isolates for the epidemiological year 2015–2016 (n = 190) available via the Meningitis Research Foundation Meningococcus Genome Library database (available at: http://pubmlst.org/perl/bigsdb/bigsdb.pl?db=pubmlst_neisseria_mrfgenomes; accessed July 2017) and (2) a subset of the isolates previously used to define the sublineages/strains of lineage 11.1 by core genome analysis [12, 15] (n = 60; Supplementary Table 2 and the Neisseria PubMLST database [available at: https://pubmlst.org/bigsdb?db=pubmlst_neisseria_isolates; accessed July 2017]).
RESULTS
Features of Sequenced Carriage Genomes
After de novo assembly, the 125-bp paired Illumina reads from carriage isolates produced contiguous sequences between 2018737 and 2155185 bp in size, consistent with expectations for meningococcal genomes (Supplementary Table 1). Genome assemblies were automatically annotated in a gene-by-gene approach by using the BIGSdb platform, and strain designation data were extracted (Supplementary Table 1). For comparison, identical typing information was extracted from the WGS data of all invasive United Kingdom MenW (n = 200) and MenY (n = 104) isolates recovered during the same epidemiological year (2015–2016) and available via the Meningococcus Genome Library database (Supplementary Tables 3 and 4, respectively).
For MenW, isolates from cc11 predominated (95%), with the strain designation W: P1.5,2: F1-1: ST-11 (cc11) accounting for 88% of cases of MenW carriage and 72% of MenW invasive isolates, respectively (Table 1). Breakdown by isolation time point confirmed that isolates with the W: P1.5,2: F1-1: ST-11 (cc11) designation were responsible for the increase in MenW carriage detected during the 2015–2016 carriage study (Table 1).
Table 1.
Breakdown of MenW Carriage and Invasive Isolates, by Strain Designation, 11.1 Sublineage, and Strain Type and by Isolation Time Point
| Strain Designation,a 11.1 Sublineage,b Strain Typeb | Isolation Time Point | Total Carriage Isolates (n = 49) |
Invasive Isolates, 2015–2016 (n = 200) |
Total Carriage and Invasive Isolates (n = 249) |
||
|---|---|---|---|---|---|---|
| Sep 2015 (n = 5) |
Nov 2015 (n = 20) |
Mar 2016 (n = 24) |
||||
| W: P1.5,2: F1-1: ST-11 (cc11) | ||||||
| South American–United Kingdom | ||||||
| 2013 | 2 | 16 | 19 | 37 | 101 | 138 |
| Original | 2 | 0 | 0 | 2 | 40 | 42 |
| South American | 0 | 2 | 2 | 4 | 0 | 4 |
| Hajj | ||||||
| Not determined | 0 | 0 | 0 | 0 | 2 | 2 |
| W: P1.5,2: F1-1: ST-ND (cc11) | ||||||
| South American–United Kingdom | ||||||
| 2013 | 0 | 0 | 1 | 1 | 15 | 16 |
| Original | 0 | 0 | 0 | 0 | 6 | 6 |
| W: P1.5,2: F1-1: ST-10651 (cc11) | ||||||
| South American–United Kingdom | ||||||
| Original | 1 | 1 | 0 | 2 | 8 | 10 |
| W: P1.5,2: F1-146: ST-11 (cc11) | ||||||
| South American–United Kingdom | ||||||
| Original | 0 | 0 | 0 | 0 | 8 | 8 |
| W: P1.5,2: F1-146: ST-ND (cc11) | ||||||
| South American–United Kingdom | ||||||
| Original | 0 | 0 | 0 | 0 | 2 | 2 |
| Other cc11c | ||||||
| South American–United Kingdom | ||||||
| 2013 | 0 | 0 | 0 | 0 | 1 | 1 |
| Original | 0 | 0 | 0 | 0 | 7 | 7 |
| Other non-cc11d | ||||||
| Not applicable | ||||||
| Not applicable | 0 | 1 | 2 | 3 | 10 | 13 |
Abbreviations: cc, clonal complex; ST, sequence type.
Derived from genome sequence data.
As assigned by core genome analysis (shown in Figure 1).
Includes all cc11 strain designations occurring only once.
Includes all non-cc11 strain designations.
For MenY, isolates from cc23 predominated (87%; Table 2). Despite the overall number of MenY isolates being smaller than that of MenW isolates, a greater diversity was evident, with MenY populations encompassing a larger number of unique strain designations than MenW (39 vs 26). Breakdown by isolation time point revealed the sporadic recovery of isolates from different MenY designations during the carriage study, with only 1 designation, Y: P1.5-1,10-1: F4-1: ST-12176 (cc23), detected at all isolation time points.
Table 2.
Frequency of Strain Designations in the MenY Carriage and Invasive Collections
| cc,a Strain Designationa | Isolation Time Point | Total Carriage Isolates (n = 32) |
Invasive Isolates, 2015–2016 (n = 104) |
Total Carriage and Invasive Isolates (n = 136) | ||
|---|---|---|---|---|---|---|
| Sep 2015 (n = 14) |
Nov 2015 (n = 8) |
Mar 2016 (n = 10) |
||||
| cc23 | ||||||
| Y: P1.5-1,10-1: F4-1: ST-1655 (cc23) | 4 | 0 | 1 | 5 | 40 | 45 |
| Y: P1.5-1,10-4: F4-1: ST-23 (cc23) | 2 | 3 | 0 | 5 | 7 | 12 |
| Y: P1.5-1,10-1: F4-1: ST-ND (cc23) | 0 | 0 | 0 | 0 | 12 | 12 |
| Y: P1.5-2,10-1: F4-1: ST-23 (cc23) | 0 | 0 | 0 | 0 | 9 | 9 |
| Y: P1.5-1,10-4: F4-1: ST-1655 (cc23) | 1 | 0 | 2 | 3 | 4 | 7 |
| Y: P1.5-1,10-1: F4-1: ST-12176 (cc23) | 1 | 1 | 2 | 4 | 1 | 5 |
| Y: P1.5-1,10-4: F4-1: ST-ND (cc23) | 0 | 0 | 0 | 0 | 5 | 5 |
| Y: P1.5-1,10-1: F4-1: ST-11754 (cc23) | 0 | 0 | 0 | 0 | 3 | 3 |
| Y: P1.5-1,10-10: F4-1: ST-1655 (cc23) | 2 | 0 | 0 | 2 | 0 | 2 |
| Y: P1.5-1,10-8: F4-1: ST-1655 (cc23) | 1 | 0 | 1 | 2 | 0 | 2 |
| Otherb | 1 | 2 | 1 | 4 | 12 | 16 |
| Non-cc23 | ||||||
| Y: P1.21,16: F3-7: ST-1466 (cc174) | 0 | 0 | 0 | 0 | 3 | 3 |
| Y: P1.18-7,9: F3-9: ST-ND (cc103) | 0 | 1 | 1 | 2 | 0 | 2 |
| Y: P1.5-1,10-4: F3-4: ST-10730 (cc167) | 0 | 0 | 0 | 0 | 2 | 2 |
| Y: P1.5-1,10-22: F5-1: ST-ND (cc22) | 0 | 0 | 0 | 0 | 2 | 2 |
| Otherc | 2 | 1 | 2 | 5 | 4 | 9 |
Abbreviations: cc, clonal complex; ST, sequence type.
Derived from genome sequence data.
Includes all cc23 strain designations occurring only once.
Includes all non-cc23 strain designations occurring only once.
WGS Analysis Resolves MenW:cc11 Carriage Isolates to the 2013-Strain of the South American–United Kingdom Sublineage
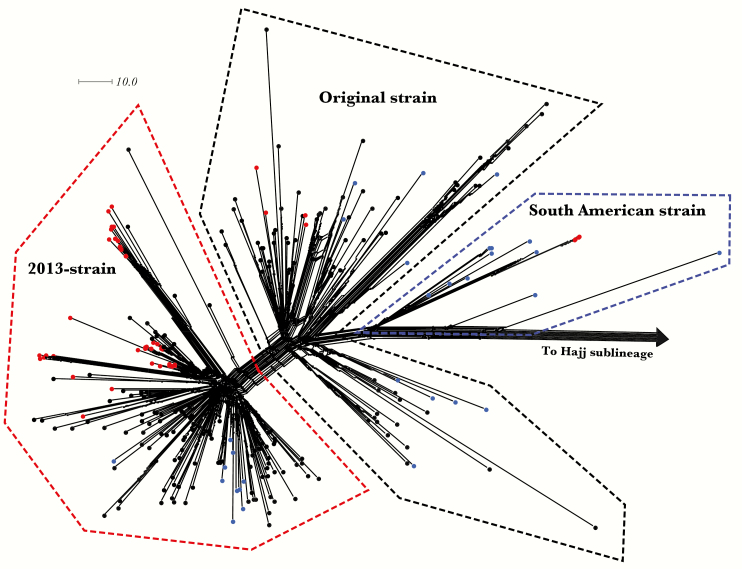
Higher-resolution genealogical analysis of the MenW:cc11 isolates was realized by comparing the core genome sequences of carriage and invasive MenW:cc11 isolates (n = 236). Identification of lineage 11.1 sublineages and strains was facilitated by inclusion of 60 additional isolates previously assigned on the basis of findings of core genome analysis [12, 15]. Carriage and invasive MenW:cc11 isolates predominantly resolved to the 2013-strain cluster within the South American–United Kingdom sublineage (83% and 62%, respectively; Figure 1 and Table 1). Core WGS analysis resolved isolates sharing the predominant W: P1.5,2: F1-1: ST-11 (cc11) designation to different 11.1 sublineages (ie, South American–United Kingdom and Hajj) and, within the South America–United Kingdom sublineage, into different strain types (Table 1). Isolation time point analysis revealed that although isolates with the W: P1.5,2: F1-1: ST-11 (cc11) designation from the original and South American strains were recovered at multiple time points, the increase in MenW carriage was almost entirely due to W: P1.5,2: F1-1: ST-11 (cc11) isolates from the 2013-strain (2, 16, and 19 isolates recovered during September, November, and March, respectively; Table 1).
Figure 1.
NeighborNet network based on the comparison of 1605 core genome loci among lineage 11.1 genomes (n = 296). Three sets of isolates were included: (1) MenW:cc11 United Kingdom carriage isolates (n = 46), (2) MenW:cc11 United Kingdom 2015–2016 invasive isolates (n = 190), and (3) previously assigned MenW:cc11 isolates (n = 60). Thirty-eight carriage isolates localized to the 2013-strain of the South American–United Kingdom sublineage, whereas 4 carriage isolates resolved to each of the original and South American strain clusters, respectively. Thirty previously assigned isolates and 2 MenW:cc11 United Kingdom 2015–2016 invasive isolates resolved to the Hajj sublineage. Nodes are color-coded, with carriage isolates in red, invasive isolates in black, and previously assigned isolates in blue. The scale bar denotes the number of allelic differences. This figure is available in black and white in print and in color online.
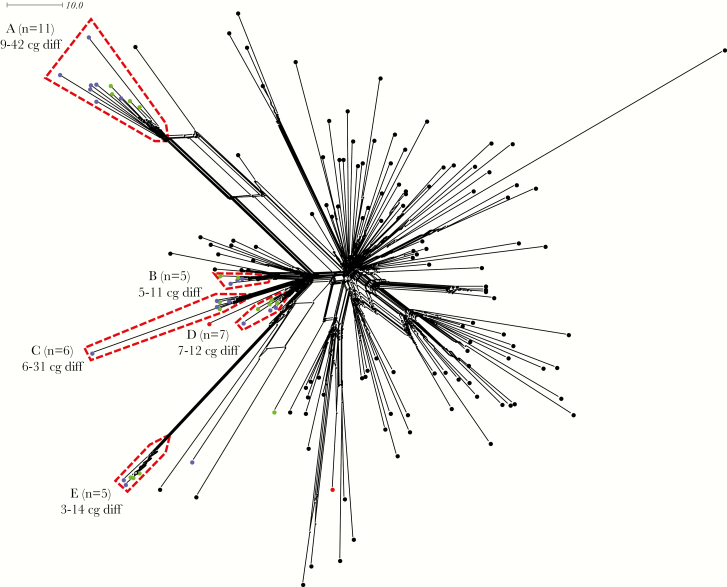
To visualize the relationships among isolates from the 2013-strain type more clearly, a NeighborNet network was generated from a separate core genome comparison of the carried (n = 38) and invasive (n = 117) 2013-strain isolates, with color-coding of nodes detailing provenance. This revealed a multifocal expansion of carriage isolates, with 97% of the isolates from November 2015 and March 2016 resolved to 5 clusters (Figure 2). Isolates within each cluster differed at only a small number of core genome loci, suggesting multiple close transmission networks. The MenACWY coverage rate for clusters A–E ranged from 57% to 83%, confirming that transmission within all 5 networks was not restricted to unvaccinated individuals (Figure 2). As sampling of students in November 2015 and March 2016 occurred in 5 dormitories [25], we examined whether 2013-strain isolate clusters correlated with dormitory of isolation. Each cluster contained isolates recovered from at least 3 different sampling sites, suggesting that transmission networks extended beyond individual dormitories (Supplementary Figure 1).
Figure 2.
NeighborNet network based on the comparison of 1605 core genome loci among 2013-strain isolates. Two sets of isolates were included: (1) 2013-strain MenW:cc11 United Kingdom carriage isolates (n = 38) and (2) 2013-strain MenW:cc11 United Kingdom 2015–2016 invasive isolates (n = 117). Nodes are color-coded, with invasive isolates in black, September 2015 carriage isolates in red, November 2015 carriage isolates in green, and March 2016 carriage isolates in blue. A total of 97% of the November 2015 and March 2016 carriage isolates resolved to 5 clusters (A–E), with isolates within each cluster being highly similar, as revealed by the number of core genome differences (cg diff). A total of 82%, 60%, 83%, 57%, and 80% of carriers harboring the isolates in clusters A–E, respectively, had received MenACWY before or during registration (September 2015). The scale bar denotes the number of allelic differences. This figure is available in black and white in print and in color online.
Effect of MenACWY Vaccination on Group W and Y Capsule Expression
Mucosal immune responses elicited by MenACWY have the potential to influence capsule expression in target serogroups. We investigated whether circulating MenW and MenY isolates shifted toward a nonserogroupable (ie, acapsulate) phenotype, but no significant changes were detected in the proportions of MenW:cc11 original strain, MenW:cc11 2013-strain, other MenW, MenY:cc23, or other MenY isolates expressing capsule between time points (Table 3). Likewise, the specific incidences of encapsulated or acapsulate MenY:cc23 (or other MenY) isolates in the population showed no significant changes during the study. For MenW isolates, the increasing incidence of carriage was driven by significant increases in both encapsulated and noncapsulated MenW:cc11 2013-strain isolates (Table 3). Notably 85% of MenW:cc11 2013-strain isolates recovered in March 2016 expressed capsule, with 82% of individuals carrying the encapsulated 2013-strain isolates at this time point having received MenACWY before or during September 2015 (Table 3).
Table 3.
Prevalence of Capsule-Expressing and Acapsulate MenW and MenY Genogroups, by Isolation Time Point
| Time (Participants, No.), Capsule Expression Status | MenW:cc11 2013-Strain Only | MenW:cc11 Original Strain Only | Other MenW | MenY:cc23 Only | Other MenYa | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Isolates, No. (%) | Participants, % (95% CI) | Isolates, No. (%) | Participants, % (95% CI) | Isolates, No. (%) | Participants, % (95% CI) | Isolates, No. (%) | Participants, % (95% CI) | Isolates, No. (%) | Participants, % (95% CI) | |
| Sep (769) | ||||||||||
| On | 0 | 0 | 2 (67) | 0.3 (0.0–0.6) | 0 | 0 | 8 (67) | 1.0 (0.3–1.8) | 1 (50) | 0.1 (0.0–0.4) |
| Off | 2 (100) | 0.3 (0.0–0.6) | 1 (33) | 0.1 (0.0–0.4) | 0 | 0 | 4 (33) | 0.5 (0.0–0.1) | 1 (50) | 0.1 (0.0–0.4) |
| Nov (353) | ||||||||||
| On | 9b (56) | 2.5 (0.9–4.2)c | 1 (100) | 0.3 (0.0–0.8) | 2 (67) | 0.6 (0.0–1.3) | 1 (17) | 0.3 (0.0–0.8) | 2 (100) | 0.6 (0.0–1.3) |
| Off | 7d (44) | 2.0 (0.5–3.4)e | 0 | 0 | 1 (33) | 0.3 (0.0–0.8) | 5 (83) | 1.4 (0.2–2.6) | 0 | 0 |
| Mar (268) | ||||||||||
| On | 17f (85) | 6.3 (3.4–9.3)g | 0 | 0 | 0 | 0 | 4 (57) | 1.5 (0.0–2.9) | 0 | 0 |
| Off | 3h (15) | 1.1 (0.0–2.4) | 0 | 0 | 4 (100) | 1.5 (0.0–2.9) | 3 (43) | 1.1 (0.0–2.4) | 2 (100) | 0.7 (0.0–1.8) |
Abbreviations: cc, clonal complex; CI, confidence interval.
Serogrouping data were unavailable for 1 MenY:cc103 isolate from March 2016.
Five (56%) were from participants who received MenACWY before or during registration (September 2015).
P < .0001, compared with the preceding time point.
Six (86%) were from participants who received MenACWY before or during registration (September 2015).
P < .01, compared with the preceding time point.
Fourteen (82%) were from participants who received MenACWY before or during registration (September 2015).
P < .05, compared with the preceding time point.
All 3 were from participants who received MenACWY before or during registration (September 2015).
Absence of Isolates From cc11 and cc23 Among Nongroupable Carriage Isolates
We obtained WGS data for an additional 93 nongroupable carriage isolates (ie, isolates lacking ctrA or carrying the capsule null locus). Extracted strain designations showed that isolates from cc198 were most prevalent (35%), followed by those from cc53 (26%) and those from cc865 (9%). Importantly, no isolates from the relevant IMD-associated MenW cc and MenY cc (cc11 and cc23, respectively) were detected, suggesting that deletion of part or all of the capsule locus by isolates of these lineages, to avoid vaccine-induced immune responses, had not occurred.
DISCUSSION
Since its appearance in 2013, cases of IMD in the UK due to the 2013-strain of the South American-UK sub-lineage of MenW:cc11 have approximately doubled year-on-year, while expansion of the original strain has slowed [15]. Here we show that the increase in carried MenW detected at a United Kingdom university during 2015–2016 [25] was also due to expansion of the 2013-strain, a finding that was reliant on the ability of core genome analysis to resolve apparently indistinguishable isolates sharing the designation W: P1.5,2: F1-1: ST-11 (cc11). Importantly, both the original and the 2013-strain MenW:cc11 strains were carried by incoming students, yet only the 2013-strain expanded. This suggests differences in strain transmissibility and/or host susceptibility to oropharyngeal carriage in this population. Further studies may indicate whether the differential expansion relates to the previously determined 4 point mutations and 3 distinct recombination events that distinguish the strains [15] or other, as yet undetermined genetic differences. The 2013-strain is a highly virulent strain with a notable tendency for atypical clinical presentation and a high case-fatality rate [13–15], and our findings suggest that it may also result in relatively high levels of carriage in semiclosed communities of young adults, a phenomenon not previously detected for the original strain or MenC:cc11 [32]. Such settings may act as a reservoir for the 2013-strain, leading to case clusters or outbreaks of disease in susceptible students [22] and onward transmission to unvaccinated cohorts in the wider population.
Of concern, the expansion of the 2013-strain occurred in the context of a student population that had, for the most part, received conjugate MenACWY. This vaccination had been introduced specifically because of the rapid and sustained increase in MenW:cc11 IMD in the United Kingdom [16] and led to a reduction in MenW cases in the first vaccine-targeted United Kingdom cohort that entered university [33]. Vaccination targeted older adolescent and young adults to provide direct protection to these age groups but also to generate indirect herd protection, as observed for the MenC and MenA monovalent conjugate vaccines, in which high vaccine coverage in these age groups reduced serogroup-specific carriage [32, 34]. Evidence supporting a comparable impact of MenACWY on carriage is currently lacking, although 2 studies in different populations showed that MenACWY elicited a modest impact on meningococcal carriage in vaccinated individuals [35, 36]. In a study involving United Kingdom university students, carriage rates of serogroup Y and combined serogroups CWY were significantly lower 2 months after MenACWY receipt [35], whereas in Polish soldiers meningococcal carriage was 9.6% in unvaccinated individuals and 1.2% in individuals who received MenACWY 1–3 years previously [36]. Of note, however, prior to vaccination serogroup Y carriage predominated over serogroup C and W carriage in the former study, and serogroup Y and C carriage were dominant in the latter study, suggesting that the observed effects of MenACWY on carriage were predominantly due to reductions in carriage of serogroup Y or carriage of Y and C strains, respectively.
Our data suggest that the MenW component of MenACWY does not have a significant impact on MenW carriage or does so at a lower level as compared to the MenY component. Thus, the sporadic and limited recovery of MenY designations, particularly cc23 isolates, during this carriage study is indicative of an absence of transmission events in this cohort. This is in marked contrast to the findings of previous studies of meningococcal carriage in university students, where MenY strains of similar clonal complexes expanded significantly and persisted in unvaccinated populations [27, 28, 37, 38]. Furthermore, the finding that the vast majority of the isolates of the 2013-strain were expressing the W capsule at the March time point is consistent with the hypothesis that the capsular polysaccharide antigen was not under significant selective pressure from the introduction of MenACWY in this population. In contrast, in a study examining the impact of MenC monovalent conjugate vaccination on carriage, Maiden et al detected a significant reduction in both the prevalence of MenC:cc11 and the proportion of recovered MenC:cc11 isolates expressing capsule (81% in 1999 and 43% in 2001, respectively) [32]. A caveat is that the majority of MenW:cc11 transmission events may have occurred in students immunized in September and during the early part of the academic year, a period known to coincide with rapid meningococcal transmission and carriage acquisition in first-year students [19]. Thus, vaccine-elicited immune responses may have developed too slowly to influence the acquisition of MenW:cc11 but not MenY:cc23 strains. Vaccinating adolescents earlier and achieving higher coverage (ie, the aim of the routine school-based program targeting adolescents, in which preliminary coverage was >77% [39]) may reduce MenW:cc11 acquisition and carriage and eventually lead to population-wide herd immunity. Ongoing surveillance will be needed to establish whether MenW:cc11 carriage declines as these cohorts enter the university population.
Our cross-sectional study precluded comment on the duration of carriage of the 2013-MenW:cc11 strains in individuals. However, prolonged carriage of these strains in MenACWY recipients could be critically important, owing to the potential for spread to nonvaccinated individuals in the population. A longitudinal study is required to determine whether there are differences in MenW/Y carriage duration in vaccinated individuals, as implied by our current study. Additionally, an examination of the impact of MenACWY on the density of meningococcal carriage is required. In a recent study, Finn et al used quantitative PCR to assess the density of meningococcal carriage and observed temporal and individual variation of several orders of magnitude [40]. Our study cannot exclude the possibility of an effect of MenACWY on carriage density in vaccinated as compared to unvaccinated individuals.
In conclusion, we show that the hypervirulent 2013-strain of the South American–United Kingdom MenW:cc11 sublineage was responsible for an increase in group W carriage reported at a United Kingdom university. Analysis of WGS data revealed close transmission networks that extended beyond individual dormitories. Furthermore, on-campus MenACWY delivery did not prevent expansion of capsule-expressing isolates from the 2013-strain of MenW:cc11. These findings are important for predicting the rate of development of population-wide herd immunity in the United Kingdom and for protecting older unvaccinated cohorts. From January through March 2017, there were 51 cases of MenW IMD in individuals aged >45 years in England [39]. Finally, further studies are required to determine whether carriage of the 2013-strain is increasing in the wider population of older adolescents and young adults in United Kingdom and in other countries where there are similar increases in IMD due to this emerging strain.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Ray Borrow, Steve Gray, and Anthony Carr (Meningococcal Reference Unit, Public Health England, Manchester, United Kingdom) for serogrouping meningococcal isolates.
This publication made use of the Meningitis Research Foundation Meningococcus Genome Library (available at: http://www.meningitis.org/research/genome), developed by Public Health England, the Wellcome Trust Sanger Institute, and the University of Oxford as a collaboration. The project is funded by Meningitis Research Foundation.
Disclaimer.
The funders had no involvement in the design of the study or the preparation of the article.
Financial support.
This work was supported by the University of Nottingham; the Medical Research Council, United Kingdom (grant MR/M020193/1 to C. D. B.); and the Wellcome Trust (grant 098051).
Potential conflicts of interest.
J. P. has been a consultant to Specific Technologies and has received research funding from Pfizer. C. D. B. collaborates with GlaxoSmithKline. D. P. J. T. has received support from Novartis Vaccines (now a part of GlaxoSmithKline), Sanofi Pasteur, and GlaxoSmithKline, including honoraria, grants, and travel assistance for conferences. N. J. O. and L. R. G. report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Stephens DS. Biology and pathogenesis of the evolutionarily successful, obligate human bacterium Neisseria meningitidis. Vaccine 2009; 27 (Suppl 2):B71–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harrison LH, Trotter CL, Ramsay ME. Global epidemiology of meningococcal disease. Vaccine 2009; 27 (Suppl 2):B51–63. [DOI] [PubMed] [Google Scholar]
- 3. Caugant DA, Maiden MC. Meningococcal carriage and disease–population biology and evolution. Vaccine 2009; 27 (Suppl 2):B64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weber MV, Claus H, Maiden MC, Frosch M, Vogel U. Genetic mechanisms for loss of encapsulation in polysialyltransferase-gene-positive meningococci isolated from healthy carriers. Int J Med Microbiol 2006; 296:475–84. [DOI] [PubMed] [Google Scholar]
- 5. Crum-Cianflone N, Sullivan E. Meningococcal vaccinations. Infect Dis Ther 2016; 5:89–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maiden MC, Jansen van Rensburg MJ, Bray JE, et al. . MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 2013; 11:728–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. von Gottberg A, du Plessis M, Cohen C, et al. . Emergence of endemic serogroup W135 meningococcal disease associated with a high mortality rate in South Africa. Clin Infect Dis 2008; 46:377–86. [DOI] [PubMed] [Google Scholar]
- 8. Abad R, López EL, Debbag R, Vázquez JA. Serogroup W meningococcal disease: global spread and current affect on the Southern Cone in Latin America. Epidemiol Infect 2014; 142:2461–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ladhani SN, Beebeejaun K, Lucidarme J, et al. . Increase in endemic Neisseria meningitidis capsular group W sequence type 11 complex associated with severe invasive disease in England and Wales. Clin Infect Dis 2015; 60:578–85. [DOI] [PubMed] [Google Scholar]
- 10. Lahra MM, Enriquez RP; National Neisseria Network. Australian meningococcal surveillance programme annual report, 2015. Commun Dis Intell Q Rep 2016; 40:E503–E11. [PubMed] [Google Scholar]
- 11. Mustapha MM, Marsh JW, Harrison LH. Global epidemiology of capsular group W meningococcal disease (1970–2015): Multifocal emergence and persistence of hypervirulent sequence type (ST)-11 clonal complex. Vaccine 2016; 34:1515–23. [DOI] [PubMed] [Google Scholar]
- 12. Lucidarme J, Hill DM, Bratcher HB, et al. . Genomic resolution of an aggressive, widespread, diverse and expanding meningococcal serogroup B, C and W lineage. J Infect 2015; 71:544–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Campbell H, Parikh SR, Borrow R, Kaczmarski E, Ramsay ME, Ladhani SN. Presentation with gastrointestinal symptoms and high case fatality associated with group W meningococcal disease (MenW) in teenagers, England, July 2015 to January 2016. Euro Surveill 2016; 21:30175. [DOI] [PubMed] [Google Scholar]
- 14. Moreno G, Lopez D, Vergara N, Gallegos D, Advis MF, Loayza S. [Clinical characterization of cases with meningococcal disease by W135 group in Chile, 2012]. Rev Chilena Infectol 2013; 30:350–60. [DOI] [PubMed] [Google Scholar]
- 15. Lucidarme J, Scott KJ, Ure R, et al. . An international invasive meningococcal disease outbreak due to a novel and rapidly expanding serogroup W strain, Scotland and Sweden, July to August 2015. Euro Surveill 2016; 21:30395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Campbell H, Saliba V, Borrow R, Ramsay M, Ladhani SN. Targeted vaccination of teenagers following continued rapid endemic expansion of a single meningococcal group W clone (sequence type 11 clonal complex), United Kingdom 2015. Euro Surveill 2015; 20:21188. [DOI] [PubMed] [Google Scholar]
- 17. Christensen H, May M, Bowen L, Hickman M, Trotter CL. Meningococcal carriage by age: a systematic review and meta-analysis. Lancet Infect Dis 2010; 10:853–61. [DOI] [PubMed] [Google Scholar]
- 18. Soriano-Gabarró M, Wolter J, Hogea C, Vyse A. Carriage of Neisseria meningitidis in Europe: a review of studies undertaken in the region. Expert Rev Anti Infect Ther 2011; 9:761–74. [DOI] [PubMed] [Google Scholar]
- 19. Neal KR, Nguyen-Van-Tam JS, Jeffrey N, et al. . Changing carriage rate of Neisseria meningitidis among university students during the first week of term: cross sectional study. BMJ 2000; 320:846–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Biswas HH, Han GS, Wendorf K, et al. . Notes from the Field: Outbreak of serogroup B meningococcal disease at a University - California, 2016. MMWR Morb Mortal Wkly Rep 2016; 65:520–1. [DOI] [PubMed] [Google Scholar]
- 21. Soeters HM, McNamara LA, Whaley M, et al. ; Centers for Disease Control (CDC). Serogroup B meningococcal disease outbreak and carriage evaluation at a college - Rhode Island, 2015. MMWR Morb Mortal Wkly Rep 2015; 64:606–7. [PMC free article] [PubMed] [Google Scholar]
- 22. Bassi C, Taha M, Merle C, et al. . A cluster of invasive meningococcal disease (IMD) caused by Neisseria meningitidis serogroup W among university students, France, February to May 2017. Euro Surveill 2017; 22:30574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Trotter CL, Maiden MC. Meningococcal vaccines and herd immunity: lessons learned from serogroup C conjugate vaccination programs. Expert Rev Vaccines 2009; 8:851–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Turner DP, Oldfield NJ, Bayliss CD. University vaccine campaign increases meningococcal ACWY vaccine coverage. Public Health 2017; 145:1–3. [DOI] [PubMed] [Google Scholar]
- 25. Oldfield NJ, Cayrou C, AlJannat MAK, et al. . Rise in Group W Meningococcal Carriage in University Students, United Kingdom. Emerg Infect Dis 2017; 23:1009–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ala’Aldeen DA, Neal KR, Ait-Tahar K, et al. . Dynamics of meningococcal long-term carriage among university students and their implications for mass vaccination. J Clin Microbiol 2000; 38:2311–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bidmos FA, Neal KR, Oldfield NJ, Turner DPJ, Ala’Aldeen DAA, Bayliss CD. Rapid clonal expansion, persistence and clonal replacement of meningococcal carriage isolates in a 2008 university student cohort. J Clin Microbiol 2011; 49:506–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ala’Aldeen DAA, Oldfield NJ, Bidmos FA, et al. . Carriage of meningococci by university students, United Kingdom. Emerg Infect Dis 2011; 17:1761–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bankevich A, Nurk S, Antipov D, et al. . SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bratcher HB, Corton C, Jolley KA, Parkhill J, Maiden MC. A gene-by-gene population genomics platform: de novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes. BMC Genomics 2014; 15:1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maiden MC, Ibarz-Pavón AB, Urwin R, et al. . Impact of meningococcal serogroup C conjugate vaccines on carriage and herd immunity. J Infect Dis 2008; 197:737–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Campbell H, Edelstein M, Andrews N, Borrow R, Ramsay M, Ladhani S. Emergency meningococcal ACWY vaccination program for teenagers to control group W meningococcal disease, England, 2015–2016. Emerg Infect Dis 2017; 23:1184–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kristiansen PA, Diomandé F, Ba AK, et al. . Impact of the serogroup A meningococcal conjugate vaccine, MenAfriVac, on carriage and herd immunity. Clin Infect Dis 2013; 56:354–63. [DOI] [PubMed] [Google Scholar]
- 35. Read RC, Baxter D, Chadwick DR, et al. . Effect of a quadrivalent meningococcal ACWY glycoconjugate or a serogroup B meningococcal vaccine on meningococcal carriage: an observer-blind, phase 3 randomised clinical trial. Lancet 2014; 384:2123–31. [DOI] [PubMed] [Google Scholar]
- 36. Korzeniewski K, Skoczyńska A, Guzek A, et al. . Effectiveness of immunoprophylaxis in suppressing carriage of Neisseria meningitidis in the military environment. Adv Exp Med Biol 2015; 836:19–28. [DOI] [PubMed] [Google Scholar]
- 37. Oldfield NJ, Harrison OB, Bayliss CD, Maiden MC, Ala’Aldeen DA, Turner DP. Genomic analysis of serogroup Y Neisseria meningitidis Isolates reveals extensive similarities between carriage-associated and disease-associated organisms. J Infect Dis 2016; 213:1777–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deasy AM, Guccione E, Dale AP, et al. . Nasal Inoculation of the Commensal Neisseria lactamica Inhibits Carriage of Neisseria meningitidis by Young Adults: A Controlled Human Infection Study. Clin Infect Dis 2015; 60: 1512–20. [DOI] [PubMed] [Google Scholar]
- 39. Public Health England. Health Protect Rep 2017; 11(23). https://www.gov.uk/government/publications/health- protection-report-volume-11-2017/hpr-volume-11-issue-23-news-30-june. Accessed September 2017. [Google Scholar]
- 40. Finn A, Morales-Aza B, Sikora P, et al. . Density distribution of pharyngeal carriage of meningococcus in healthy young adults: New approaches to studying the epidemiology of colonization and vaccine indirect effects. Pediatr Infect Dis J 2016; 35:1080–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.