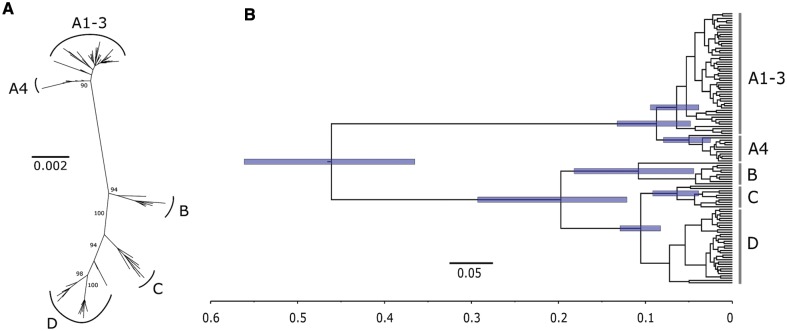
Fig. 1.
Phylogenetic tree of the 118 HPV16 complete genomes. (A) Unrooted ML phylogenetic tree inferred using the complete coding region alignment after excluding a total of 26 positively selected and 115 negatively selected codons. The final alignment encompassed 118 sequences, 6,093 bp and 316 distinct alignment patterns. Bootstrap support after 500 replicates is given for branches leading to the A1–3, A4, B, C, and D variant lineages. Scale bar in substitutions per nucleotide site. (B) Maximum clade credibility tree inferred for selection-filtered HPV16 genome coding region alignment under the HHS scenario, 9.6 × 10−9 [5.5 × 10−9–13.6 × 10−9] subs/site/yr as substitution rate prior and enforcing two time point calibrations: archaic Hominin divergence at 500 kya (95% CI, 400–600 kya) and recent modern human migration out-of-Africa at 90 kya (95% CI, 60–120 kya). Scale bar in millions of years ago. Node bars indicate the 95% probability intervals for the corresponding node age.