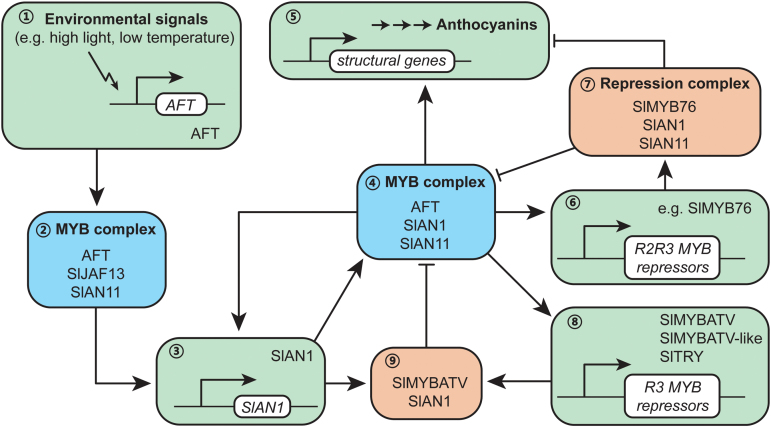
Fig. 7.
A putative model of the anthocyanin gene regulation network in the peel of tomato fruit. This model is modified from fig. 8 in the previously published paper [Albert et al. (2014). A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. The Plant Cell 26, 962–980. Republished with permission (www.plantcell.org), ‘Copyright American Society of Plant Biologists’] according to the expression patterns and putative functions of the corresponding genes that we determined in the present study. In the fruit peel of Aft/Aft (abbreviation of Aft/Aft ATV/ATV) plants (1) anthocyanin biosynthesis is initiated by activating the expression of the AFT gene under high light or low temperature. (2) The AFT protein (MYB activator, most likely SlAN2 or SlAN2-like) interacts with SlJAF13 and SlAN11 to form a MBW complex that activates the expression of SlAN1 (3). (4) SlAN1 interacts with AFT and SlAN11 to form a core MBW activation complex, and the complex activates the expression of the SlAN1 gene and most of the structural genes (5), and enhances the anthocyanin pigmentation. (6) The MBW complex also activates the expression of R2R3 MYB repressors. (7) The R2R3 MYB repressor, most likely SlMYB76, might intrude into the MBW complex and repress the anthocyanin pigmentation. (8) The MBW complex also activates the expression of R3 MYB repressors. (9) The R2R3 MYB repressor, e.g. SlMYBATV, competes with AFT to bind SlAN1 to inhibit the formation of new MBW complexes. This feedback inhibition can prevent the production of too much anthocyanin.