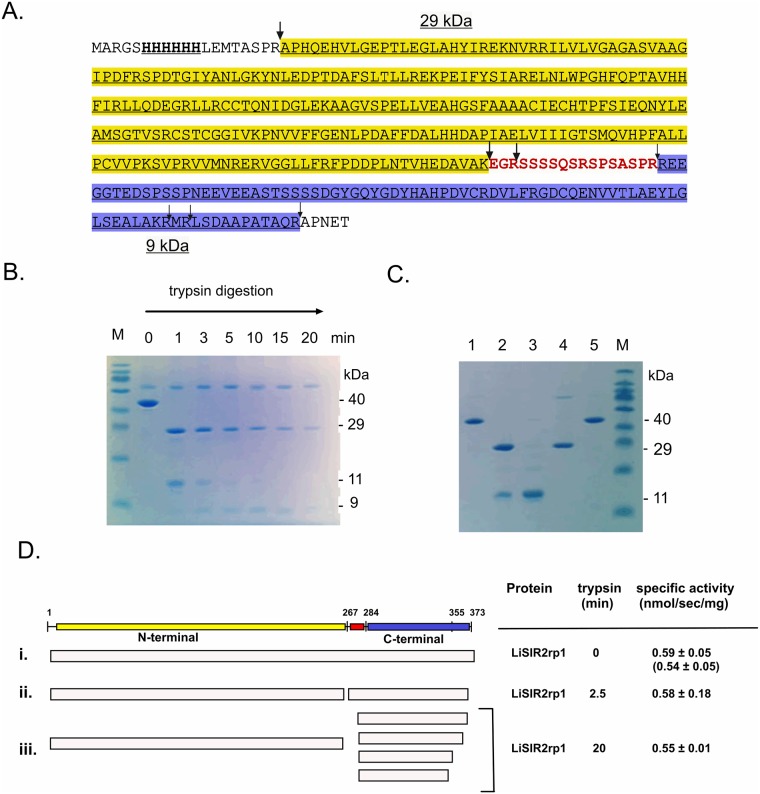
Fig 1. Probing the proteolytically-stable domain structure of LiSIR2rp1 and its deacetylase activity.
(A) Primary amino acid sequence of 6 x His N-terminal tagged LiSIR2rp1 showing positions sensitive to tryptic digestion under non-denaturing conditions (arrows), defined from SDS PAGE and LC/ESI-TOF MS (see Methods and S1 Text). The larger arrows indicate principal digestion sites in time-resolved proteolysis. The proteolytically-sensitive region linking proteolytically-stable N-terminal (yellow) and C-terminal (blue) regions is shown in red. (B) Time-resolved tryptic digestion (0–20 min) of LiSIR2rp1 showing appearance of an N-terminal, ~29 kDa region (28580.83 and 28921.53 Da) and a ~ 9 kDa C-terminal region (9593.98, 9081.4, 7999.25 and 7710.43 Da) derived from an initial ~ 11 kDa digestion product (t = 1 to 3 min). (The larger protein at 66 kDa is albumin in the quenching buffer). (C) Re-purification of His tagged LiSIR2rp1 after native tryptic digestion; a 6 x His LiSIR2rp1 ΔN5-373 mutant protein lacking the N-terminal trypsin site was used for these experiments (see Methods). Lanes 1 and 5, undigested LiSIR2rp1 purified on Ni2+- affinity column. Lane 2, native digest of LiSIR2rp1 re-purified on Ni2+-affinity column (N- and C- termini bind). Lane 3, 6 M urea wash of native digest of LiSIR2rp1 bound to Ni2+- affinity column (C-terminal domain removed). Lane 4, Elution of native digest from Ni2+-affinity column with imidazole after 6 M urea wash (N-terminus alone). (D) Sirtuin deacetylase activity of LiSIR2rp1 after native tryptic digestion. Time resolved digests of 6 x His LiSIR2rp1 ΔN5-373 were re-purified from trypsin reactions by Ni2+- affinity chromatography before assessing specific activity on the p53 acetyl lysine substrate. Specific activity (nmol deacetylated product/sec/mg protein) was compared for (i) undigested protein (t = 0) measured both for 6xhis LiSIR2rp1 ΔN5-373 and full length protein (shown in parentheses), (ii) a 2.5 min digest and (iii) a 20 min digest. The fragmentation patterns were deduced from the results of SDS PAGE and LC/MS analyses.