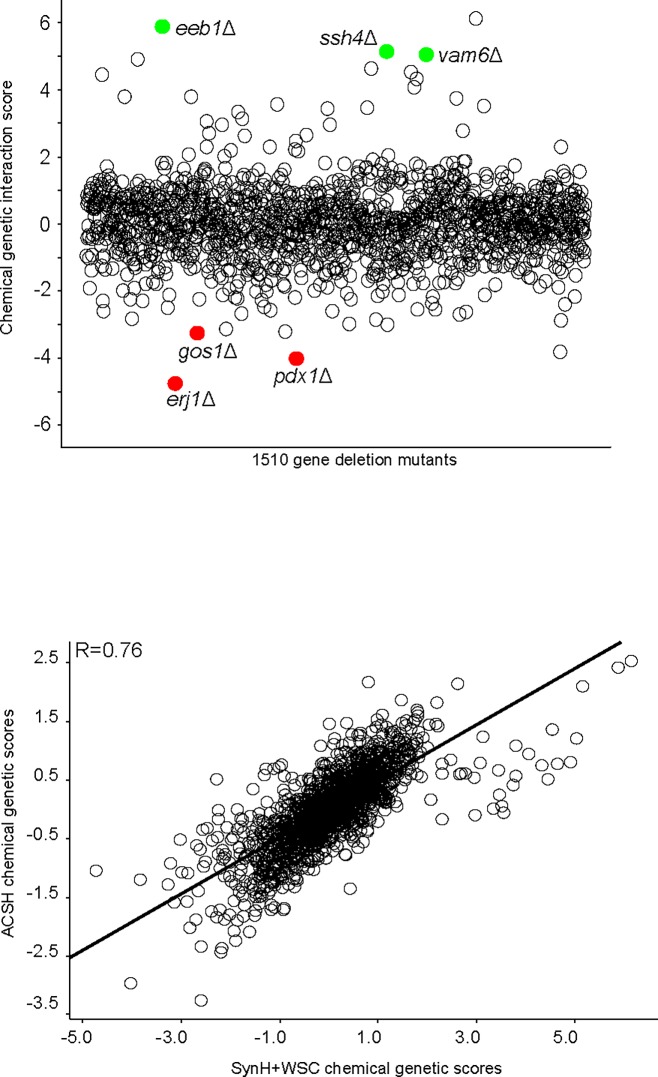
Fig 6. Chemical genomic profiling of WSC on using a yeast deletion strain library.
(A). Chemical genomic profile of SynH+WSC, with the top sensitive deletion mutants (red) and top resistant mutants (green) highlighted (mean profile n = 3); (B). Gene clusters correlation between SynH, ACSH and SynH+WSC (n = 3). Mutants in ERJ1 (involved in ER protein folding), PDX1 (a subunit of the mitochondrial dehydrogenase complex), and GOS1 (involved Golgi transport) were especially sensitive to WSC. The sensitive genes gave insight into the mechanism of toxicity, confirming that cell membranes were the likely target of WSC toxicity; and overexpression of the sensitive genes could be used to confer resistance. Comparing the chemical genetic profiles between different hydrolysates and fermentation media, we found that the profile of SynH + WSC exhibited a high correlation with that of ACSH (Fig 6B, R = 0.69), and showed greater similarity to ACSH compared to SynH (Fig 7). The strong correlation suggests that the degradation compounds in WSC can represent the real inhibitors in ACSH to a large extent.