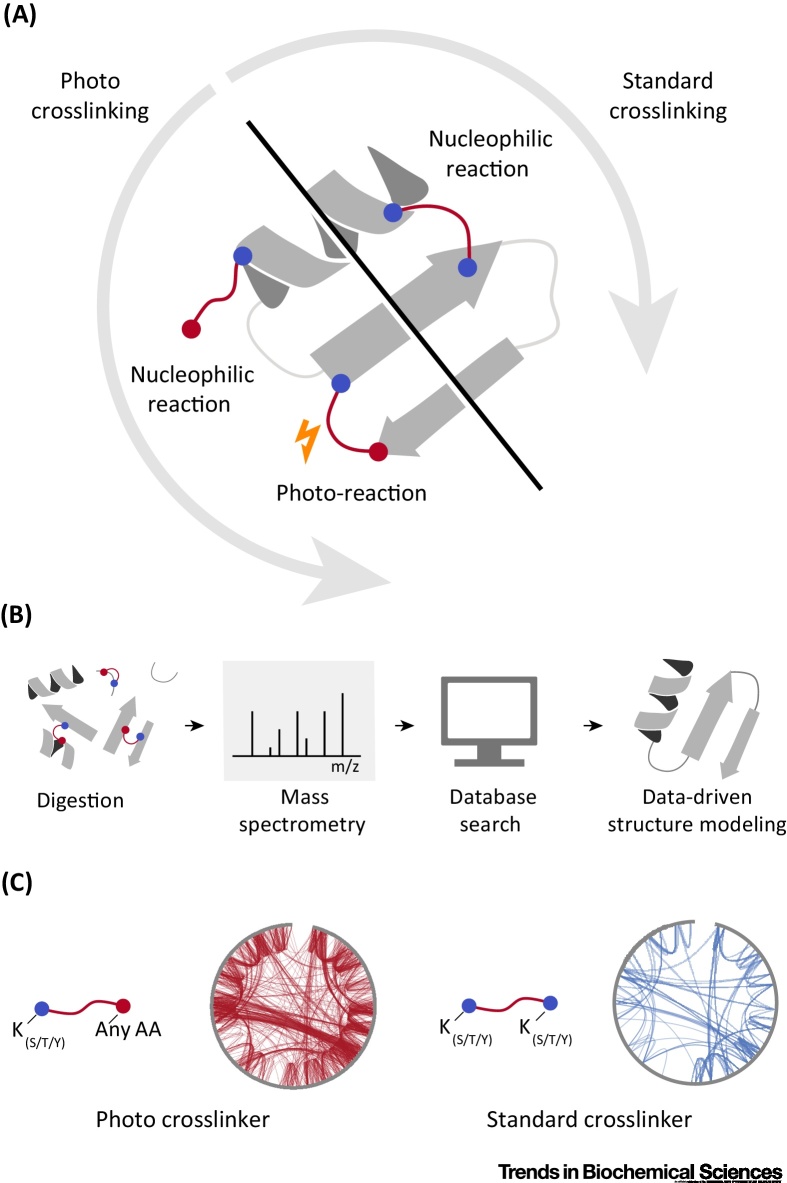
Figure 1.
Overview of a Crosslinking Experiment for Protein Structure Determination. (A) As the first step in standard (homobifunctional) crosslinking, the crosslinker reacts with a specific reactive residue and then a second one to form a crosslink. Photo-crosslinking with photoactivatable reagents follows the same workflow. However, in the nucleophilic reaction step, only one side of the crosslinker reacts with the protein. The other side is activated by UV light and then reacts with the protein to form the crosslink. (B) The experimenter digests the protein using proteases (usually trypsin). The resulting peptides are then subjected to mass spectrometry. Specialized database search software reads out the crosslinks from the mass spectrometry data. The crosslinks then form the input to data-driven protein structure modeling. (C) Photo-crosslinkers such as sulfosuccinimidyl 4,4′-azipentanoate react on one side with lysine (and S/T/Y) and can react with any amino acid on the other side. This leads to a high crosslink density (the sequence of the protein is depicted by the circle; the crosslinks are shown as lines). These crosslinks can be leveraged for structural modeling. The reaction specificity of standard homobifunctional crosslinkers targets lysines (and S/T/Y residues to a lesser extent). This limits the density of the resulting crosslink network.