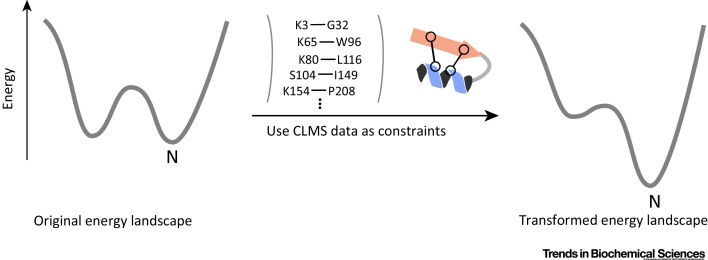
Figure 2.
Effect of Crosslinking/Mass Spectrometry (CLMS) Data in Conformational Space Search. De novo protein structure modeling searches the conformational space of the protein for the lowest energy conformation, which usually coincides with the native structure. However, the energy landscape is rugged, and the energy of the native state might be close to the energy of other local minima. This makes search difficult because there might be no clear gradient toward the native structure. Using CLMS data as residue–residue constraints transforms the energy landscape by deepening the energy well of the native structure. This also makes the energy landscape less rugged and provides a gradient toward the native state. This makes it easier to search for the native conformation and therefore leads to more frequent sampling of nativelike structures in de novo structure modeling calculations.