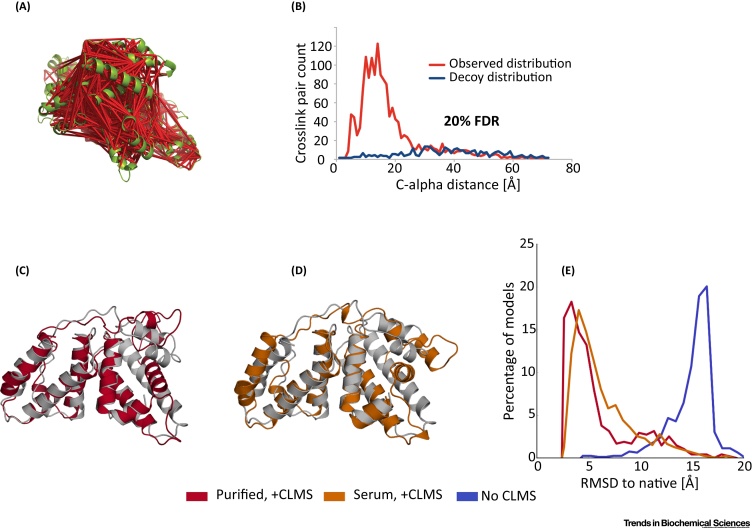
Figure 3.
Using Photo-Crosslinking/Mass Spectrometry (CLMS) Crosslinkers for Structure Modeling. (A) Photo-crosslinking of human serum albumin (HSA) with sulfosuccinimidyl 4,4′-azipentanoate leads to 1495 links at 20% false-discovery rate. (B) The distance distribution of crosslinked residues follows a log-normal distribution. Most crosslinks are between residues with Cα distances below 20 Å. (C) The combination of high-density CLMS data with computational protein modeling is able to recapitulate the HSA domain structures. Here, we show the results for domain C of HSA. Models are shown in color, while the native structure is shown in gray. Using high density-CLMS (HD-CLMS) data from purified HSA samples leads to modeled structures with a root mean square deviation (RMSD) of 2.9 Å. (D) Using HD-CLMS data from HSA samples in blood serum leads to models with an RMSD of 3.8 Å to the native structure. (E) RMSD distribution of low-energy computed models using CLMS data from purified HSA (red), from HSA in blood serum (orange), and without CLMS data (blue). Using CLMS data shifts the RMSD distribution toward lower RMSD values. Thus, the CLMS effectively guides conformational space search and allows to sample nativelike, low-RMSD structures more frequently. Adapted from [18].