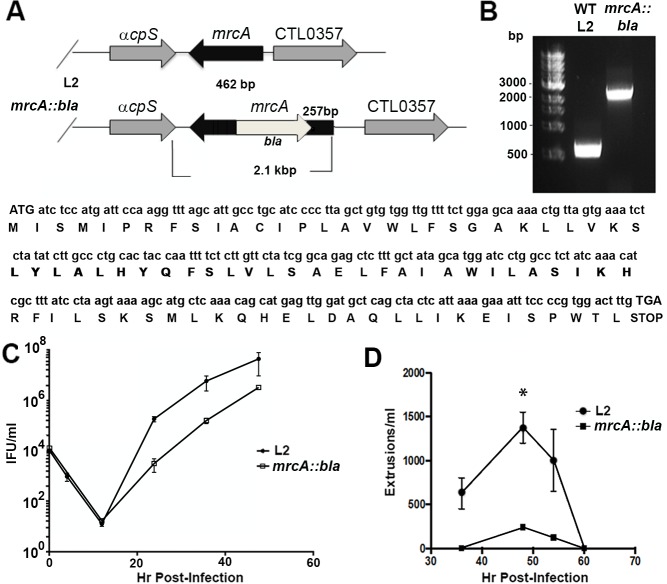
Fig 2. Generation of an mrcA::bla mutant.
(A) Sanger sequencing of isolated DNA using gene-specific primers was conducted to verify orientation and insertion of the intron. (B) PCR products were separated on a 1% agarose gel, DNA was visualized using ethidium bromide staining. WT, Wild type. (C) Hela cells infected with wild-type L2 or mrcA::bla L2 were incubated for 0 h, 4 h, 12 h, 24 h, 36 h, or 48h. Cells were lysed in water and replated on fresh monolayers to enumerate the progeny IFUs. Experiments were performed in triplicate (n = 3, error bars indicate SEM. (D) Hela cells infected with wild-type L2 or mrcA::bla L2 at an MOI = 1. At various time points post infection, the supernatants were removed and gently pelleted by centrifugation at 1200rpm. The resulting pellet was resuspended in 100 μl of media. Extruded inclusions free of host cell nuclei were enumerated (n = 3, error bars represent SEM). The difference was significant (P<0.01) by an unpaired Students T-test at 48hpi.