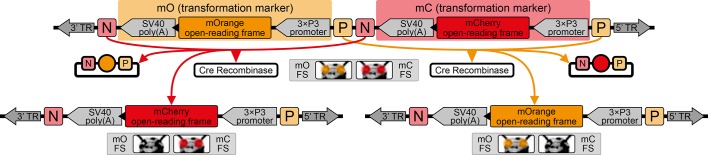
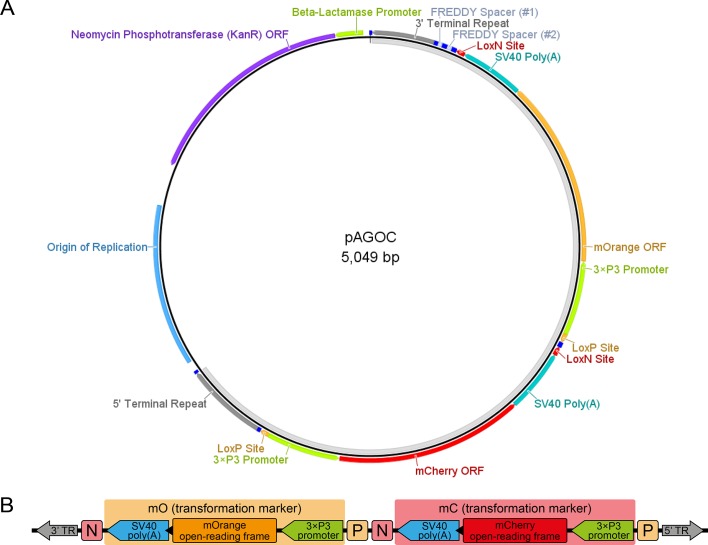
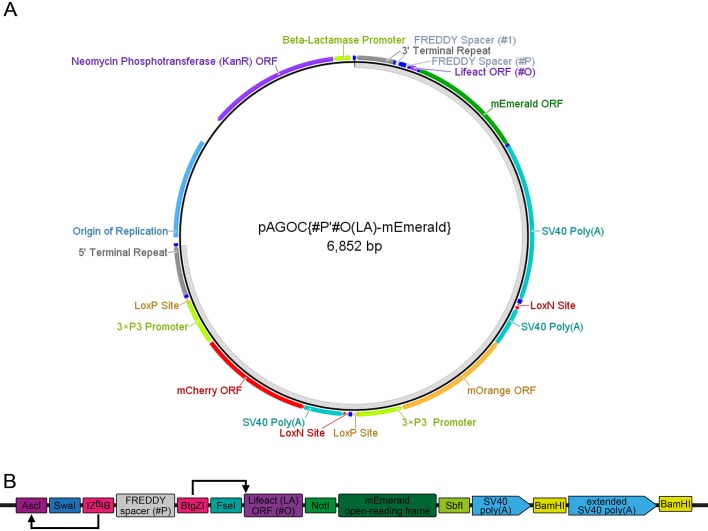
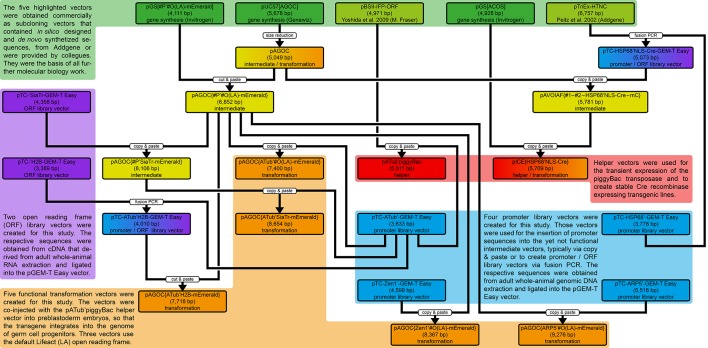
Figure 1. The AGameOfClones vector concept within the piggyBac-based transformation-ready pAGOC vector for Tribolium.
Two fluorescence-based transformation markers, mO and mC, are embedded into a piggyBac-based transformation-ready vector, which is characterized by 3’ and 5’ terminal repeats (TR) necessary for genomic insertion. The markers are based on the artificial eye-specific 3×P3 promoter, the open-reading frame for the respective fluorescent protein, that is, mOrange or mCherry, and the SV40 poly(A). Each transformation marker is flanked upstream by a LoxP site (P) and downstream by a LoxN site (N), forming interweaved Lox site pairs. The markers can be detected in the eyes by using appropriate filter sets (FS). Cre-mediated recombination leads to the excision of one marker from the genome. Upon removal, the other marker remains within the genome, since the remaining LoxP and LoxN sites are incompatible. Individuals that underwent recombination give rise to progeny in which only one marker is detected in the eyes.