Abstract
Some longstanding dogmas in the circadian field warrant reexamination in light of recent studies focused on the role of post-translational modifications and intrinsic disorder in core circadian clock proteins of mice and fungi. Such dogmas include the role of turnover in circadian feedback loops and the origin myths describing evolutionary relatedness among circadian clocks. In this Essay, the authors recapitulate recent findings on circadian clock protein regulation by taking an unconventional approach in the form of a dialog between Wizard and Apprentice.
They sat quietly, and after a while the Apprentice said to the Wizard, “Tell me a just-so story, you know, one that is easy to remember because everything fits together so well in the end.”
“Well,” smiled the Wizard, “I guess if it’s a just-so story, it doesn’t need to be true, but just sound like it might have been true, so here is one.” And the Wizard spoke with closed eyes.
“Once upon a time, there was an animal cell, or a fungus, with a circadian clock, and the clock worked this way. Two proteins made a heterodimer that acted as a transcription factor (TF). The TF drove expression of a gene or genes encoding proteins that made a complex with other proteins; this complex physically interacted with the heterodimeric TF and reduced the TF’s activity so that fewer of the repressive proteins were made. In the fullness of time, important proteins in the repressive complex were post-translationally modified, especially by phosphorylation. And we all know what phosphorylation does, don’t we? It leads to protein turnover. So the important proteins in the repressive complex were degraded. This released the TF to start a new cycle, and the clock ran along happily ever after.”
“It’s a wonderful story,” the Apprentice mused sadly, “but I guess if it’s a just-so story, then it isn’t really all true, is it?”
“That’s right,” volunteered the Wizard, “it’s not all true. But there’s no reason to be sad! This just-so story has been a circadian dogma for a long time, and while some parts are true, the truth about the other parts is still emerging—and is more interesting than a just-so story, anyway. And as you will one day be a wizard too, you may want to know this.” And so the Wizard continued.
“There really is a daily cycle of activation and repression of a heterodimeric TF made of two different proteins; they can bind to DNA, and they interact via PAS domains. In mammals, these are BMAL1 and CLOCK (BCC), and in Neurospora, they are WC-1 and WC-2 (WCC). In each case, they drive expression of proteins (PERs and CRYs in mammals; FRQ in Neurospora) that enter into complexes with other proteins and then physically interact with BCC/WCC to depress the TF’s activity. PERs, CRYs, and FRQ are phosphorylated and turned over, typically just before the reactivation of BCC/WCC in the daily cycle. The just-so story part is not about what happens, but about why it happens: recent experiments are making it look a lot like it’s not the turnover, but instead the phosphorylation of FRQ and CRY, that releases the WCC/BCC from repression, and that turnover is just an afterthought—good cellular housekeeping, but in no way a deterministic step in the normal daily circadian cycle.
“Of course,” said the Wizard, warming up to the topic and to a captive audience, “there are ways to build a clock that do not involve transcription-translation feedback loops or protein turnover at all. Cyanobacteria build a clock that can run in a test tube with just three proteins and two phosphorylation sites (Nakajima et al., 2005), and that advances through a wonderfully choreographed sequence of subtle and sometimes rare and transient structural changes that eventually lead back to just where they started (Abe et al., 2015), and that …”
As the Wizard gulped for air, the Apprentice gently queried, “Just-so stories.?”
“Ah, yes; I digress. Just-so stories, indeed. Anyway, as I was saying, clues had been building that the dogma was incomplete (for review, see Hurley et al., 2016; Larrondo et al., 2015). The initial impetus to revising the dogma came from Neurospora, the tractable and durable model for clocks in animals and fungi, where, for instance, the molecular basis of light-resetting was first understood. The dogma makes some unambiguous predictions, among them that period length and protein stability are mechanistically linked rather than merely correlated and that the cycle cannot continue while there are high levels of the repressive proteins. Neither prediction, however, holds up.
“The backstory here is that although phosphorylation of a protein can lead to its turnover, it can also change a protein’s activity and influence its interacting partners. Indeed, for some repressive clock proteins it appears that both activity and turnover are influenced by phosphorylation. It’s been known for decades that the negative-element clock proteins are rhythmically phosphorylated. For instance, FRQ is heavily phosphorylated in a time-of-day-specific manner, and some of the phosphorylations lead to turnover (Baker et al. 2009). In wild-type cells, there is an excellent correlation between stability and period length, and similarly good correlations are seen between PER stability and period length in the mechanistically similar oscillator in Drosophila (Syed et al., 2011) and between Cry1 half-life and period length in mammals when CRY is engineered to be less stable (Ode et al., 2017). These universal correlations are part of what made this just-so story a dogma.
“The surprise came when strains of Neurospora were deleted for FWD-1, the F-box protein that targets phosphorylated FRQ for degradation. Although these deletion strains were overtly arrhythmic based on loss of circadian developmental cycles, just as the dogma predicts, when the oscillator itself was directly followed through frq-luciferase transcriptional or FRQ-luciferase translational fusion reporters, it was seen to run along smoothly even in the presence of un-degraded, highly phosphorylated FRQ (Larrondo et al., 2015). More thorough analyses confirmed that there was no causative link between FRQ half-life and period length: although phosphorylation can influence turnover as well as activity, in normal cells it appears that the effect on activity is what controls the cycle length. To conceptualize this, Larrondo et al. (2015) divided the FRQ phosphorylation sites into two classes. One class, the clock-signaling phosphorylations (CSPs), contributed to timing and happened in a predictable processive manner, leading in the end to a protein that is inactive in the cycle by virtue of its phosphorylation status. The other class was the termination-signaling phosphorylations (TSPs) that led to protein turnover. FWD-1 is signaled through the TSPs, but in a normal cycle CSPs precede TSPs, and CSPs determine the length of the cycle. Plainly, though, if TSPs were engineered into FRQ so they happened too soon, they would lead to changes in FRQ levels that could impact period determination.
“Of course,” opined the Wizard, “some dismissed the Neurospora conclusions because Neurospora is not an animal and FRQ bears little sequence conservation with CRY or PER. But when others looked deeper into this story, the parallels were more compelling. Recently, an elegant study from the Ueda lab examined similar issues in the mouse (Ode et al., 2017). The study is a technical tour-de-force, revealing an embryonic stem cell methodology whereby gene-rescue experiments in whole mice can be executed in a generation. They turned this tool on the question of circadian period control by CRY1, one of the salient repressive proteins of the mammalian oscillator, and in so doing they tested the dogma that clock protein turnover was inextricably linked to period determination.”
“Ah,” volunteered the Apprentice, “I remember now. CRY1 and its paralog CRY2 cycle both in abundance and phosphorylation status, and the clock can run without either one, but not without both (van der Horst et al., 1999; Vitaterna et al., 1999). So they could make Cry1/Cry2 double-knockout cells whose clock would be completely dependent on whatever CRY they reintroduced. That’s clever.”
“Yes, that’s it exactly,” said the Wizard, continuing. “Ode et al. mapped and mutated the phosphorylation sites on CRY1 and then asked (1) whether non-phosphorylatable mutants could rescue rhythms in mouse embryonic fibroblasts (MEFs) lacking any CRY and (2) whether the mutations affected CRY1 stability. The mutants that could not rescue rhythms in the Cry1/Cry2 double-knockout MEFs were further subdivided based on the response of the clock to constitutive overexpression of the proteins: overexpression of some of the CRY mutants conferred a dominant period shortening, some resulted in hyper-repression and arrhythmicity, and some resulted in hypo-repression, as if the construct was invisible or wasn’t even there. Placing these alleles on the CRY1 structure drew attention to a blob on Cry1, the region around the highly flexible P loop that interacts with PER2, and to the fact that period effects in double mutants were plainly additive in a manner reminiscent of double mutants in FRQ phosphosites (Baker et al., 2009). When they considered only those proteins whose stability was affected by the mutation, they were able to show a weak correlation between stability and period length. Through use of a clever method, they were able to show that changes in stability could influence period length, within limits. This is like engineering TSP into a clock protein. But this period-altering effect held only for those proteins whose stability was affected by the mutation. The more surprising observation was that when all the mutants were examined, there were a number that displayed drastic period alterations without significant stability changes, demonstrating that there was no obligate correlation between period length and stability. Stated differently, this means that changes in CRY1 stability are sufficient, but not necessary, to effect period changes, a result fully compatible with the undogmatic conclusions of Larrondo et al. (2015) from FRQ and Neurospora.
“The interpretation of these data is that when, through genetic manipulations, TSPs happen too soon, they can lead to changes in CRY levels that will impact period determination, but this is not the normal case. In this landmark study, Ode et al. used the CRY1 mutants to generate (in one generation) mice whose sole source of this essential circadian repressor was the phosphosite mutant Cry1, and the analysis of these strains is definitive. Some CRY1 phosphorylations must happen for the clock to run; for instance, CRY1(S261A) mice are arrhythmic. If some phosphorylations happen too soon (as mimicked by Ser-to-Asp), the cycle is short-circuited and runs fast (e.g., CRY1[S243D] and others), or in some cases the clock never can run at all because the mutant CRY1, though stable, is invisible to the clock (CRY1 [S261D] or CRY1[Y432D]). Indeed, in this way the study provides textbook examples of CSPs: sites that, when they are phosphorylated, inactivate the repressive function. These must be phosphorylated for the clock to run because their modification effectively closes the feedback loop and releases the heterodimeric TF to start a new cycle, but if they are constitutively phosphorylated, they yield proteins that are as stable as or even more stable than wild-type CRY1 but that, even when overexpressed, are invisible to the clock.”
“Invisible proteins with highly flexible or disordered blobs–wow!” cried the Apprentice.
“OK, not exactly invisible,” winced the Wizard, self-correcting, “but rather just ineffectual blobs. But let’s talk more about highly flexible and/or intrinsically disordered (ID) regions, as there is an emerging story there also.
“We are taught as Apprentices that proteins are supposed to function because they have structures, but many proteins, in particular parts of proteins involved in signaling, regulation, and transcriptional activation, are ID (for review, see Wright and Dyson, 2015). An emerging consensus is that this is particularly so with core proteins of circadian clocks. If you recall, the core clock proteins include the positive elements BMAL1 and CLOCK in mammals and their corresponding components in fungi, WC-1 and WC-2, as well as the negative elements PER and CRY and the corresponding part in fungi, FRQ. Major portions of all of these proteins are ID.
“The first demonstration of this was for the C-terminal domain of human CRY2 (Partch et al., 2005), where this region was suggested to be important for signaling. WC-1 and WC-2 are predicted to have large regions of disorder in their C termini and putative transcriptional activation domains (TADs), consistent with the appearance of ID regions within the TADs of transcription factors (e.g., Wells et al., 2008). Additionally, FRQ has been shown by a variety of methods to be an ID protein that must be stabilized by another protein, a so-called ‘Nanny,’ in order for it to function in the clock (Hurley et al., 2013). Circular dichroism demonstrated ID regions in the C-terminal signaling regions of CRY and BMAL (Czarna et al., 2011), and more recently Xu et al. demonstrated the importance of disorder in the TAD of BMAL1 for interaction with CRY1 (Xu et al., 2015). In this study, the Partch lab showed that CRY1 is able to repress BMAL1 transactivation by competing with CBP (p300) for binding to the intrinsically unstructured C-terminal TAD of BMAL1, with binding eliciting dynamic conformational rearrangements. We can perhaps add to this list the highly flexible P loop and surrounding regions of CRY1 studied by the Ueda lab, whose phosphorylation is central to period determination as described above (Ode et al., 2017). Yet, despite all this disorder, circadian proteins do form complexes, implying inherent structure; the mammalian PERs, CRYs, CLOCK, BMAL1, and CK1δ are all present in a 30-polypeptide, 1.9-MDa complex seen during the repressive phase of the clock cycle (Aryal et al., 2017).
“Proof of significant ID in circadian proteins has used a variety of approaches ranging from structural prediction programs such as PONDR, to biochemical methods (e.g., freeze-thaw sensitivity or protease sensitivity), to sophisticated biophysical methods (like circular dichroism), to appealingly simple methods like solubility after boiling. You’ll like this method,” said the Wizard, with a wink. “Proteins with structure remain in solution because the hydrophobic residues are inside and the hydrophilic residues are outside, all held together after careful folding. If this meticulous folding is disrupted by high temperatures, the protein falls out of solution, like a boiled egg. However, interestingly, if you boil FRQ (Hurley et al., 2013) or Per2 (J.M. Hurley, J.J.L., and J.C.D., unpublished), they stay in solution, apparently because for most of the protein there is no inside or outside to unfold. And yes, Apprentice, you can try this at home.
“We can argue from first principles several corollaries from this perhaps surprising observation of so much disorder in circadian proteins. First is the realization that ID means that signaling interactions can be more plastic than with proteins of more fixed structure. Multiple binding partners are typically seen with ID proteins (Wright and Dyson, 2015), and ID means both that there should be little energy required to transition between states, and that partners can influence shape as well as shape being able to influence partners. Because ID regions are typically surface exposed, they are common targets for chemically, spatially, and (importantly) temporally distinct post-translational modifications, and these modifications in turn can drive spatially and temporally distinct changes in activity and interactions by allowing integration of multiple weak signals. Consistent with this idea, a hallmark of circadian regulators is their highly choreographed daily cycles of post-translational modification (for review, see Reischl and Kramer, 2011). For example, BMAL1 is subject to multiple chemically, spatially, and temporally distinct modifications, and FRQ is phosphorylated at more than 100 sites in a characteristic spatially, and temporally, distinct pattern prior to its inactivation by phosphorylation (Baker et al., 2009; Larrondo et al., 2015). Here’s a generic view of this kind of circadian cycle,” declared the Wizard, and with upturned arms Figure 1 was conjured and floated in the air.
Figure 1. Related Mechanistic Features between Animal and Fungal Clocks.
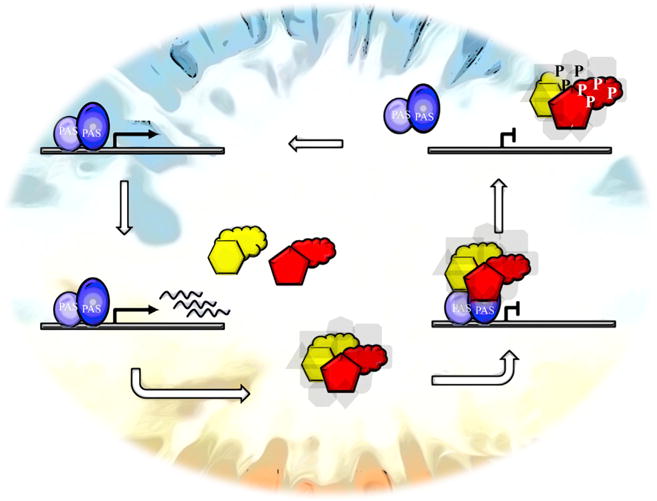
Beginning in the upper left, parts of the heterodimeric TF (oval and circle) interact via PAS domains and bind to the promoters of genes encoding negative elements. The negative-element proteins (yellow and red) have both structured (hexagon and pentagon) and flexible or unstructured regions (clouds). The negative elements form a complex in the nucleus with other proteins (gray background shapes) including CK1 and interact with the TF, causing it to be inactivated. Multisite clock-signaling phosphorylations (P) of the negative elements lead to their inactivation as repressors, and to dissociation, releasing the heterodimeric TF to restart the cycle.
“Another corollary of the disorder found within parts of circadian proteins impacts how we view their evolutionary conservation. The requirement that a protein achieve a specified structure means that the primary sequence must also be conserved, and indeed sequence elements from the highly structured core DNA-binding and PAS-PAS-interaction domains are shared between BMAL1/ CLOCK in mammals and WC-1/WC-2 in Neurospora (Lee et al., 2000). However, absent this structural constraint, primary sequences in ID regions can evolve more quickly, and partners interacting with those domains might be recruited or lost based on weak interactions and thus may not be conserved. This tendency is also well known among the repressive clock components. For instance, CRY, the principal circadian repressor in mammals and star of the story told here, has no role in the core Drosophila clock, where it acts only as a photoreceptor. Similarly, Timeless, an essential clock protein for Drosophila, appears to play at best a peripheral role in the mammalian oscillator, where a paralog is associated with DNA damage-dependent resetting (Engelen et al., 2013). Given this kind of rapid change, there is little reason to expect sequence conservation between the repressive elements of animals and fungi.”
“I guess that colors our origin myth for fungal and animal circadian clocks then, too, doesn’t it?” reasoned the Apprentice. “If you focus on the clock genes first identified in fungi and animals, all of them repressive components, you see widely diverged sequences. But if you focus instead on the activator proteins and regions that have structure, and on the post-translational modifiers and regulatory logic of the feedback loop, then the similarities are striking. In all cases, heterodimeric TFs interacting via PAS domains drive expression of their own repressors, and the long circadian time constant is regulated by post-translational modification. Looking at it this way, the evolutionary origin of these clocks looks like evolution of everything else, where animals and fungi group together (e.g., Forterre, 2015). Do I have it about right, Wizard?”
The Wizard smiled. “Yes, you do.”
References
- Abe J, Hiyama TB, Mukaiyama A, Son S, Mori T, Saito S, Osako M, Wolanin J, Yamashita E, Kondo T, Akiyama S. Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock. Science. 2015;349:312–316. doi: 10.1126/science.1261040. [DOI] [PubMed] [Google Scholar]
- Aryal RP, Kwak PB, Tamayo AG, Gebert M, Chiu PL, Walz T, Weitz CJ. Macromolecular Assemblies of the Mammalian Circadian Clock. Mol Cell. 2017;67:770–782.e6. doi: 10.1016/j.molcel.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL, Kettenbach AN, Loros JJ, Gerber SA, Dunlap JC. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol Cell. 2009;34:354–363. doi: 10.1016/j.molcel.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarna A, Breitkreuz H, Mahrenholz CC, Arens J, Strauss HM, Wolf E. Quantitative analyses of cryptochrome-mBMAL1 interactions: mechanistic insights into the transcriptional regulation of the mammalian circadian clock. J Biol Chem. 2011;286:22414–22425. doi: 10.1074/jbc.M111.244749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelen E, Janssens RC, Yagita K, Smits VA, van der Horst GT, Tamanini F. Mammalian TIMELESS is involved in period determination and DNA damage-dependent phase advancing of the circadian clock. PLoS ONE. 2013;8:e56623. doi: 10.1371/journal.pone.0056623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P. The universal tree of life: an update. Front Microbiol. 2015;6:717. doi: 10.3389/fmicb.2015.00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Larrondo LF, Loros JJ, Dunlap JC. Conserved RNA helicase FRH acts nonenzymatically to support the intrinsically disordered neurospora clock protein FRQ. Mol Cell. 2013;52:832–843. doi: 10.1016/j.molcel.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Loros JJ, Dunlap JC. Circadian Oscillators: Around the Transcription-Translation Feedback Loop and on to Output. Trends Biochem Sci. 2016;41:834–846. doi: 10.1016/j.tibs.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrondo LF, Olivares-Yañez C, Baker CL, Loros JJ, Dunlap JC. Circadian rhythms. Decoupling circadian clock protein turnover from circadian period determination. Science. 2015;347:1257277. doi: 10.1126/science.1257277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Loros JJ, Dunlap JC. Interconnected feedback loops in the Neurospora circadian system. Science. 2000;289:107–110. doi: 10.1126/science.289.5476.107. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Ode KL, Ukai H, Susaki EA, Narumi R, Matsumoto K, Hara J, Koide N, Abe T, Kanemaki MT, Kiyonari H, Ueda HR. Knockout-Rescue Embryonic Stem Cell-Derived Mouse Reveals Circadian-Period Control by Quality and Quantity of CRY1. Mol Cell. 2017;65:176–190. doi: 10.1016/j.molcel.2016.11.022. [DOI] [PubMed] [Google Scholar]
- Partch CL, Clarkson MW, Ozgür S, Lee AL, Sancar A. Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry. 2005;44:3795–3805. doi: 10.1021/bi047545g. [DOI] [PubMed] [Google Scholar]
- Reischl S, Kramer A. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett. 2011;585:1393–1399. doi: 10.1016/j.febslet.2011.02.038. [DOI] [PubMed] [Google Scholar]
- Syed S, Saez L, Young MW. Kinetics of doubletime kinase-dependent degradation of the Drosophila period protein. J Biol Chem. 2011;286:27654–27662. doi: 10.1074/jbc.M111.243618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells M, Tidow H, Rutherford TJ, Markwick P, Jensen MR, Mylonas E, Svergun DI, Blackledge M, Fersht AR. Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc Natl Acad Sci USA. 2008;105:5762–5767. doi: 10.1073/pnas.0801353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Gustafson CL, Sammons PJ, Khan SK, Parsley NC, Ramanathan C, Lee HW, Liu AC, Partch CL. Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nat Struct Mol Biol. 2015;22:476–484. doi: 10.1038/nsmb.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]


