Abstract
The current report describes a newly devised method for automatically scoring the incidence of rat hepatocyte micronuclei (MNHEP) via flow cytometry, with concurrent assessments of hepatocyte proliferation—frequency of Ki-67-positive nuclei, and the proportion of polyploid nuclei. Proof-of-concept data are provided from experiments performed with 6-week old male Crl:CD(SD) rats exposed to diethylnitrosamine (DEN) or quinoline (QUIN) for 3 or 14 consecutive days. Non-perfused liver tissue was collected 4 days after cessation of treatment in the case of 3-day studies, or 1 day after last administration in the case of 14-day studies for processing and flow cytometric analysis. In addition to livers, blood samples were collected one day after final treatment for micronucleated reticulocyte (MN-RET) measurements. Dose-dependent increases in MNHEP, Ki-67-positive nuclei, and polyploidy were observed in 3- and 14-day DEN studies. Both treatment schedules resulted in elevated %MNHEP for QUIN-exposed rats, and while cell proliferation effects were subtle, appreciable increases to normalized liver weights were observed. Whereas DEN caused markedly higher %MNHEP when exposure was extended to two weeks, QUIN-induced MNHEP were slightly increased with protracted dosing. Parallel microscopy-based MNHEP frequencies were highly correlated with flow cytometry-based measurements (4 study/aggregate R2 = 0.80). No increases in MN-RET were seen in any of the four studies. Collectively, these results suggest liver micronuclei are amenable to an automated scoring technique that provides objective analyses and higher information content relative to conventional microscopy. Additional work is needed to expand the number and types of chemicals tested, identify the most advantageous treatment schedules, and test the transferability of the method.
Keywords: micronuclei, liver, hepatocytes, genotoxicity, flow cytometry
Introduction
The most widely used approach for studying in vivo chromosome damaging potential is to expose laboratory rodents to test material and score the incidence of micronucleated cells. The test is most often conducted with hematopoietic cells, especially reticulocytes [Matter and Schmid 1971; Heddle, 1973; MacGregor et al., 1980]. Erythrocytes’ lack of a main nucleus, extruded before entering the peripheral blood compartment, makes observing and scoring micronucleated reticulocytes (MN-RET) a relatively rapid and straightforward process. Use of this cytogenetic damage marker has eclipsed in vivo chromosome aberration due to its greater technical ease, coupled with its sensitivity to aneugenic agents [Witt et al., 2008]. In fact for the last several decades the erythrocyte-based micronucleus assay has been a cornerstone of required safety testing for the registration of new pharmaceuticals and industrial chemicals [ICH, 2011; Hayashi et al., 2007].
Several groups have reported automated approaches for scoring MN-RET, including flow cytometry [Grawé et al., 1992; Dertinger et al., 2004], laser scanning cytometry [Styles et al., 2001], and image analysis [Asano et al., 1998]. With these platforms in place, MN-RET scoring is now more objective and the assay is conducted with improved intra- and inter-laboratory reproducibility [Torous et al., 2005; Dertinger et al., 2006; Chang et al., 2014]. Among the several significant benefits of automation are gains in efficiencies that have made it practical to integrate the MN-RET endpoint into repeat-dose rat toxicology studies, an important contribution to 3Rs—refinement, reduction, and replacement [Pfuhler, 2008].
While the erythrocyte-based micronucleus assay benefits from advanced scoring techniques and continues to represent a pivotal genetic toxicology endpoint, the most recent guidance documents stress the importance of considering other tissues in addition to those of the hematopoietic compartment. For instance the S2(R1) document of the International Conference on Harmonisation states: “The inclusion of a second in vivo assay in the battery is to provide assurance of lack of genotoxicity by use of a tissue that is well exposed to a drug and/or its metabolites, ” [ICH, 2011]. Also, in regard to certain pro-genotoxicants showing negative results in traditional chromosomal damage assays, it states: “…these examples likely reflect a lack of appropriate metabolic activity or lack of reactive intermediates delivered to the hematopoietic cells of the bone marrow.”
In practice, in vivo non-hematopoietic cell assays most commonly focus on the liver. It is the main organ responsible for drug metabolism, and the local concentration of genotoxic intermediates is often highest in this tissue. The in vivo comet and transgenic rodent mutation assays represent the current state-of-the-art for addressing liver-based genotoxicity assessments. However, these assays are relatively costly, technically challenging, not sensitive to aneugens, and/or not readily combined with other toxicology studies. Genetic toxicologists, including those involved in regulatory agency-required safety testing, are therefore in need of more efficient, 3Rs friendly methods for measuring in vivo DNA damage in non-hematopoietic tissues, especially the liver.
Collaborative work conducted by Japanese scientists under the auspices of the JEMS/MMS Group has been addressing the need for a liver-based cytogenetic damage assay. Most recently this body of work focused on a repeat-treatment rat liver micronucleus assay, and promising results were obtained for 6-week old rats exposed to test articles over 14 or 28 consecutive days. This important research was summarized in several publications, including an IWGT report [Uno et al., 2015a], as well as papers published by the JEMS/MMS group [Uno et al., 2015b, Hamada et al., 2015].
The present report describes a novel flow cytometric method that complements work being done in this area by addressing certain disadvantages with microscopy-based MNHEP scoring practices, especially in terms of throughput capacity and objectivity, with the further advantage of providing information about hepatocyte proliferation. The proof-of-principle studies described herein were conducted with diethylnitrosamine (DEN) and quinoline (QUIN), two chemicals that are reportedly negative in the MN-RET assay, but capable of inducing MNHEP [Hamada et al., 2015; Uno et al., 2015b]. Thus, while many hepatocarcinogens that require enzymatic bioactivation in order to form proximate genotoxic and tumorigenic metabolites are positive in the MN-RET assay, DEN and QUIN highlight the desirability of investigating hepatocytes in addition to hematopoietic cells, as some metabolites’ high reactivity and short-half life lead to tissue-specific responses [Kang et al., 2007; Reigh et al., 1996].
Materials and Methods
Reagents, Miscellaneous Supplies
DEN (CAS No. 55-18-5), QUIN (CAS No. 91-22-5), dimethyl sufloxide (CAS No. 67-68-5), and sesame oil (CAS. no. 8008-74-0) were purchased from Sigma-Aldrich, St. Louis, MO. Heat-inactivated fetal bovine serum (FBS; cat. no. 89510-186) was from VWR, Radnor, PA. Reagents used for flow cytometric MN-RET scoring (Anticoagulant Solution, Buffer Solution, DNA Stain, Anti-CD71-FITC and Anti-CD61-PE Antibodies, RNase Solution, and Malaria Biostandards) were from In Vivo MicroFlow® PLUS R Kits, Litron Laboratories, Rochester, NY. Reagents used for flow cytometric MNHEP scoring (Liver Preservation Buffer, Buffer Solution, Erythrocyte Clearing Solution, Collagenase Solution, Lysis Solution 1, Lysis Solution 2, Anti-Ki-67-eFluor® 660, DNA Stain (contains SYTOX® Green), and RNase Solution) were from Prototype In Vivo MicroFlow® PLUS RL Kits, Litron Laboratories. Falcon-brand 35 micron nylon mesh (cat. no. 352235) was from Corning, Corning, NY. Heparinized capillary tubes were purchased from Fisher Scientific, Pittsburg, PA (cat. no. 22-260-950).
Animals, Treatments
Experiments were conducted with the oversight of the University of Rochester’s Institutional Animal Care and Use Committee. Male Crl:CD(SD) rats were purchased from Charles River Laboratories, Wilmington, MA. Rodents were allowed to acclimate for approximately one week. Water and food were available ad libitum throughout the acclimation and experimental periods.
For preliminary work that focused on assessments of liver growth and hepatocyte proliferation as a function of age, 4, 6, and 8-week old rats (n = 4/group) were treated with water via oral gavage for 3 consecutive days. For this and all experiments described herein, dosing solutions were administered once per day at a volume of 10 mL/kg body weight/day. Four days after the final treatment, livers were removed and processed as described in detail below.
In the case of the DEN and QUIN studies, age at the start of treatment was 6 weeks, n = 5 - 6 per group. Six-week old rats were chosen to emulate the JEMS/MMS group’s work that has recently focused on this age [Uno et al., 2015b, Hamada et al., 2015]. Dose levels were chosen based on the literature as well as from preliminary dose-range-finding experiments. DEN was prepared in water at 10× concentrations, and aliquots were frozen at −20°C until use. On each day of administration, aliquots were thawed and added to water to prepare working solutions of 0, 1, 2, and 4 mg/mL for the 3-day study, and 0, 0.5, 1, and 2 mg/mL for the 14-day study. Administration was by oral gavage, for final dose levels of 0, 10, 20, or 40 mg/kg/day for the 3-day study, and 0, 5, 10, or 20 mg/kg/day for the 14-day study.
QUIN was prepared in sesame oil each day treatment occurred. Working solutions of 0, 7.5, 10, 12.5 mg/mL were prepared for the 3-day study, and 0, 5, 7.5, and 10 mg/mL for the 14-day study. Administration was by oral gavage, for final dose levels of 0, 75, 100, or 125 mg/kg/day for the 3-day study, and 0, 50, 75, or 100 mg/kg/day for the 14-day study.
Micronucleated Reticulocyte Assay
Peripheral blood was collected approximately 24 hrs after cessation of treatment, i.e., day 4 in the case of the 3-day studies, and day 15 of the 14-day studies. In the case of day 4 specimens, blood was obtained by nicking a lateral tail vein with a surgical blade after animals were warmed briefly under a heat lamp. Approximately 200 μL of free-flowing blood were collected directly into heparinized capillary tubes. Day 15 specimens were collected into MicroFlow PLUS-R kit-supplied Anticoagulant Solution-coated needles and syringes via heart puncture following CO2 anesthesia. Aliquots of each whole blood sample (50 μL) were transferred to tubes containing 175 μL Anticoagulant Solution, and they were maintained at room temperature for less than 3 hrs until fixation with ultracold methanol occurred as described by Torous and colleagues [2003].
MN-RET and reticulocyte (RET) frequencies were determined for day 4 and 15 blood samples via flow cytometry according to the In Vivo MicroFlow PLUS-R Kit manual, v170503 (www.litronlabs.com). These procedures have been described in detail previously [Torous et al., 2003; Dertinger et al., 2004]. MN-RET frequency measurements were based on the acquisition of approximately 20,000 high CD71-positive RET per blood sample. Instrument setup and calibration was performed using kit-supplied biological standards (P. berghei-infected blood cells) [Tometsko et al., 1993]. A BD FACSCalibur™ flow cytometer running CellQuest™ Pro v5.2 software was used for data acquisition and analysis.
Micronucleated Hepatocyte Assay
Livers were harvested 4 days after cessation of treatment for 3-day studies, or 1 day after final treatment for the 14-day studies. Rats underwent CO2 anesthesia and were immediately exsanguinated via heart puncture using 22 gauge, 1 inch needles and syringes (typically 6-9 mL blood were collected per rat). Thoracic cavities were opened to ensure death via pneumothorax, followed by excision of the entire liver. Wet liver weights were recorded, and left lateral lobes were transferred to 50 mL tubes containing ice-cold Liver Preservation Buffer (includes 10% v/v dimethyl sulfoxide added same day as use).
Left lateral lobes were cut with a razor blade to provide single sections of approximately 1 g per rat. Each was added to separate T25 flasks containing 10 mL 37 °C Buffer Solution and placed on a shaking incubator set to 37 °C and 130 rpm. After 10 minutes the solutions were removed and 10 mL fresh room temperature Buffer Solution were added for an additional 10 minutes of shaking at 37 °C. The solutions were removed and the process was repeated two times with 10 mL room temperature Erythrocyte Clearing Solution. After the second volume of Erythrocyte Clearing Solution was removed, 10 mL room temperature Collagenase Solution with 10% v/v heat-inactivated FBS were added. After 30 minutes of incubation at 37 °C with shaking, the Collagenase Solution was removed and 13 mL room temperature Buffer Solution + FBS were added. The contents of each flask were transferred to a 50 mL conical tube where the liver was cut into approximately 5 - 10 smaller, roughly equal-size pieces with scissors. The coarsely chopped liver material was vortexed (high setting) for approximately 3 minutes, after which the cell suspensions were transferred to conical 15 mL centrifuge tubes. Centrifugation occurred for 2 minutes at 200 × g. Supernatants were aspirated and cells were resuspended with 5 mL Buffer Solution + FBS. This washing step was repeated 1 to 3 times, until the supernatants were clear in appearance. Finally, the cells were resuspended in 3 mL Buffer Solution + FBS and strained through 35 micron nylon mesh.
Tubes of strained cells were stored overnight in a 4 °C refrigerator. This represented a convenient way to stagger work associated with tissue collection, sample preparation, and data acquisition. Furthermore, unexpected advantages were realized when flow cytometric analyses were delayed to the following day, or even as much as 4 days after the nylon mesh straining step described above. That is, samples held overnight or for several days at approximately 4 °C yielded consistently lower and less variable micronucleus frequencies that were found to correspond better to parallel microscopy-based values. Regarding the MNHEP scoring via microscopy, approximately 10 μL of each strained cell preparation were applied to acridine orange-coated slides prepared as described in Hayashi and colleagues [1990]. Slides were coded before analysis occurred using an Olympus BH-2 microscope (10× eyepiece, 40× objective) outfitted for fluorescence using blue light excitation via a mercury lamp. Photomicrographs showing representative acridine orange-stained hepatocytes are shown in Supplemental file S1.
On the day of flow cytometric analysis, an additional 5 mL cold Buffer + FBS were added and cells were gently resuspended with pipetting. Cells were centrifuged at 200 × g for 2 minutes, and supernatants were aspirated. Cells were resuspended with approximately 2 mL cold Buffer + FBS. For the remaining steps reagents were at room temperature and the samples were maintained at ambient temperature. A 100 μL aliquot of the cell suspension was transferred to a new tube, and 400 μL of Lysis Solution 1 were added slowly, over approximately 15 seconds. Upon addition of Lysis Solution 1, the tube was immediately vortexed (low setting) for 5 seconds. After 30 minutes, 400 μL Lysis Solution 2 were injected forcefully into each tube, which were immediately vortexed (low setting) for 5 seconds.
After equilibrating for at least 5 minutes, samples were analyzed with a FACSCanto™ II flow cytometer equipped with 488 nM and 633 nM excitation (BD Biosciences, San Jose, CA). Instrumentation settings and data acquisition/analysis were controlled with FACSDiva™ software v6.1.3 (BD Biosciences). SYTOX Green-associated fluorescence emissions were collected in the FITC channel (530/30 band-pass filter), and anti-Ki-67-eFluor® 660-associated fluorescence emissions were collected in the APC channel (660/20 band-pass filter). The flow cytometry gating strategy that was developed for this MNHEP scoring application required events to fall within each of three regions and one histogram marker before they were scored as nuclei or micronuclei (see Figure 1a-f).
Figure 1.
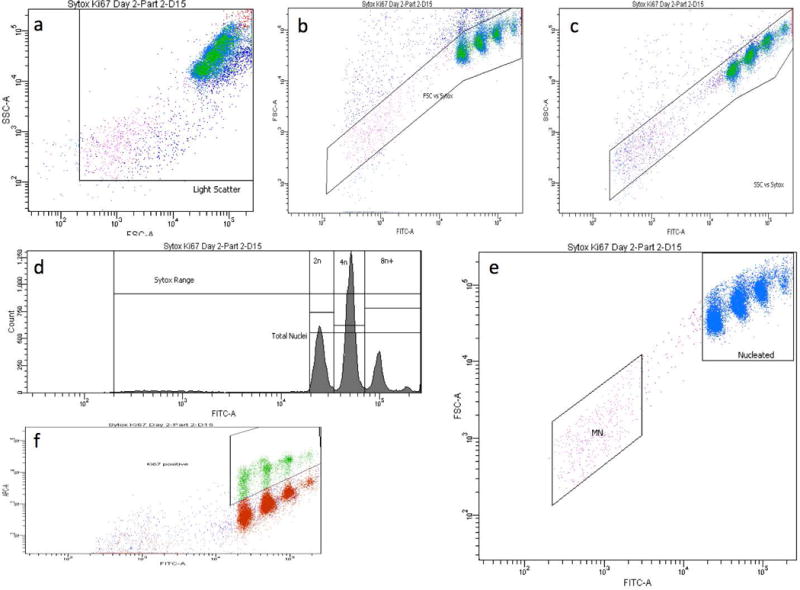
a-f. Bivariate plots and a histogram of liver nuclei and micronuclei from a rat treated with 20 mg DEN/kg/day for 14 days. These panels illustrate the gating strategy used to score the frequency of micronuclei, with simultaneous assessments of hepatocyte proliferation. In order for events to be scored as nuclei or micronuclei, they needed to meet each of the following 4 criteria: within a side scatter vs. forward scatter region, panel a; within a forward scatter vs. SYTOX Green fluorescence region, panel b; within a side scatter vs. SYTOX Green fluorescence region, panel c; and within a SYTOX Green range marker, panel d. Panel d also shows histogram markers used to enumerate the proportion of nuclei with 2n, 4n, and 8n+ ploidy status. Panel e shows the micronucleus-scoring region (MN), which was set to approximately 1/100 to 1/10 the SYTOX fluorescence intensity of G1 nuclei. Panel f shows the region used to enumerate the proportion of nuclei that exhibit anti-Ki-67-associated fluorescence.
The incidence of flow cytometry-scored MNHEP is expressed as frequency percent (no. micronuclei/no. nuclei × 100), and these values were generally based on the acquisition of 20,000 SYTOX Green-positive nuclei per specimen. Microscopy-based MNHEP frequencies were derived in the same manner, however in this case approximately 2,000 cells were evaluated per rat. Simultaneous with micronucleus assessments, several experimental indices of hepatocyte proliferation were collected. In the first case, regions were created to determine the percentage of Ki-67-positive nuclei. A second statistic used to track proliferation was the relative ratio of nuclei exhibiting 2n, 4n, and 8n and greater DNA content (8n+), determined from SYTOX Green-associated fluorescence. These ratios were converted to a single statistic that is analogous to the cytokinesis block proliferation index [Kirsch-Volders et al., 2003], and in this report it is referred to as the hepatocyte proliferation index (HPI). The HPI statistic was derived as follows: [(No. 2n nuclei) + (No. 4n nuclei *2) + (No. 8n+ nuclei *3)]/No. total nuclei. A third metric that we investigated was simply the percentage of nuclei with 8n and greater DNA content, i.e., %8n+ nuclei.
Calculations, Statistical Analyses
Each treatment group’s MNHEP and MN-RET frequencies were compared to concurrent vehicle controls using Dunnett’s multiple comparison t-tests in JMP software’s one-way analysis of variance platform (v12.0.1, SAS Institute Inc., Cary, NC). Tests were performed at the 5% level using a one-tailed test to identify significant increases relative to vehicle control. These same analyses were performed with normalized liver weights (g liver weight/kg body weight), %Ki-67-positive nuclei, HPI values, %8n+ nuclei, and %RET data as well, however in these cases the tests were two-tailed in order to identify significant increases or decreases compared to vehicle control.
Results and Discussion
Assay optimization
Early assay optimization efforts were performed with naïve rats where the emphasis was to develop liver harvest and cell processing steps that consistently provided hepatocyte preparations that almost exclusively consisted of intact, single cell suspensions that could be divided between slides for microscopic scoring, and towards detergent, dye and antibody reagents for flow cytometric analysis. This work progressed to experiments with 4-6 week old rats treated for up to three consecutive days with DEN (data not shown). Over the course of these early efforts, several important variables were identified. One key finding was that exsanguination, in conjunction with rapid removal and transfer of liver sections into cold Liver Preservation Buffer, is essential for obtaining consistent, high quality specimens. Collagenase concentration, incubation time, and temperature were also found to be crucial. Once these variables were identified and standardized, optimized Lysis Solutions with sufficient detergent/dye/antibody reagents were found to provide flow cytometric MNHEP measures that reproducibily corresponded well to parallel microscopy-based assessments.
To better understand the type of information that Ki-67, HPI, and %8n+ nuclei measurements provide as part of a micronucleus-scoring assay, an experiment was performed with groups of rats that differed in age. As shown by Figure 2a,b,c even as animals added significant body mass over a 4- week period of time, normalized liver weights and %MNHEP did not appreciably change. On the other hand, Ki-67-positive nuclei steadily decreased over this timeframe (Figure 2d). This was an expected result, as hepatocyte proliferation is known to slow as rats age [Higami et al., 1997]. Thus, this biomarker appears to provide information about the proportion of hepatocytes that recently underwent division. Conversely, HPI and %8n+ values tended to increase with rat age, although the latter did not reach statistical significance (Figure 2e,f). Since HPI and %8n+ reflect the degree of accumulated polyploidization, these indices are probably best understood as metrics that portray a longer-term view of hepatocyte division history, at least as compared to the Ki-67 biomarker. With micronucleus- and proliferation-measuring aspects of the assay in place, we proceeded to study two metabolically-activated genotoxicants in the context of two treatment/harvest schedules.
Figure 2.

a-f. Flow cytometric data for the rat age study are shown. Panel a = Change in body weight over the course of the treatment period; panel b = normalized liver weights; panel c = %MNHEP; panel d = %Ki-67-positive nuclei; panel e = hepatocyte proliferation index; panel f = %8n+ nuclei. For every graph, data for each individual rat is shown, and group means appear as green horizontal lines. Dunnett’s test results are shown to the far right of each graph, where statistically significant differences relative to the 6 week-old rat group appear as italicized black text as opposed to red text, and by grey circles as opposed to red circles. Circles’ diameters represent 95% confidence intervals.
DEN: 3-day study
While no other outward signs of toxicity were observed, body weight gains were significantly reduced after 3 consecutive days of treatment with DEN (Figure 3a). In this short-term treatment design, only the top dose group showed a modest but significant reduction to normalized liver weights (Figure 3b). Taken together, these data suggest that the dose levels used in this short-term treatment study were well tolerated.
Figure 3.
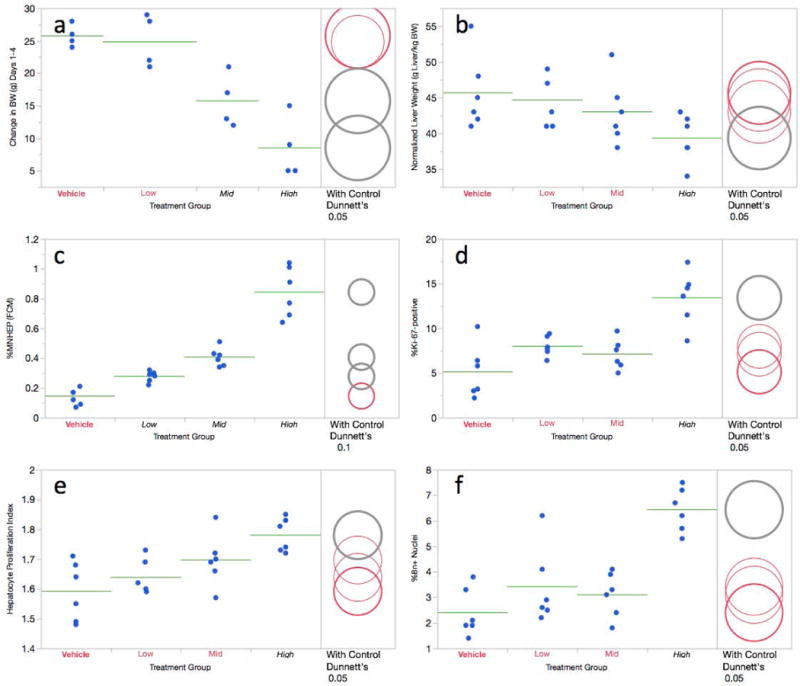
a-f. Flow cytometric data for the 3 day diethylnitrosamine study are shown. Panel a = Change in body weight over the course of the treatment period; panel b = normalized liver weights; panel c = %MNHEP; panel d = %Ki-67-positive nuclei; panel e = hepatocyte proliferation index; panel f = %8n+ nuclei. For this study, low, mid, and high treatment groups correspond to 10, 20, and 40 mg/kg/day, respectively. For every graph, data for each individual rat is shown, and group means appear as horizontal green lines. Dunnett’s test results are shown to the far right of each graph, where statistically significant differences relative to the concurrent vehicle control group appear as italicized black text as opposed to red text, and by grey circles as opposed to red circles. Circles’ diameters represent 95% confidence intervals.
As shown by Figure 3c, flow cytometry-based %MNHEP values were observed to increase in a dose-dependent manner, with each of the dose groups exhibiting significantly higher frequencies compared to vehicle control. Whereas the vehicle control rats averaged 0.14% MNHEP, animals in the top dose group exhibited a mean frequency of 0.85%, or a 6.1-fold increase.
Simultaneous with MNHEP frequencies, several measurements related to hepatocyte proliferation were collected. The frequency of Ki-67-positive nuclei showed a significant increase in the high DEN dose group (Figure 3d). Furthermore, this 40 mg/kg/day dose level also caused elevated proportions of polyploid nuclei, as evidenced by statistically significant increases to HPI and %8n+ nuclei values (Figure 3e,f).
In addition to the liver-centric analyses described above, peripheral blood cells were analyzed for RET and MN-RET frequencies. DEN exposure was found to cause modest reductions to %RET, with significant decreases observed in rats treated with 20 and 40 mg/kg/day. On the other hand, no group exhibited increased %MN-RET relative to vehicle control (Table I).
Table I.
Blood reticulocyte and micronucleated reticulocyte measurements.
| Study | Treatment (mg/kg/day) | Treatment Duration (Days) | Blood Harvest Day | %RET | %MN-RET |
|---|---|---|---|---|---|
| DEN, short-term | 0 | 3 | 4 | 4.84, 4.99, 6.23, 5.69, 6.93, 7.21 | 0.15, 0.12, 0.11, 0.19, 0.07, 0.12 |
| 10 | 3 | 4 | 5.87, 5.70, 6.00, 6.56, 5.64, 6.38 | 0.11, 0.11, 0.10, 0.08, 0.10, 0.11 | |
| 20 | 3 | 4 | 4.39, 4.92, 5.22, 4.83, 5.46, 5.60 * | 0.10, 0.06, 0.14, 0.10, 0.11, 0.14 | |
| 40 | 3 | 4 | 3.54, 3.22, 3.26, 3.32, 3.22, 4.02 * | 0.17, 0.11, 0.10, 0.12, 0.08, 0.17 | |
| DEN, protracted | 0 | 14 | 15 | 3.20, 3.30, 2.81, 3.56, 2.69 | 0.11, 0.07, 0.18, 0.05, 0.12 |
| 5 | 14 | 15 | 2.02, 3.64, 2.65, 2.55, 1.53 | 0.14, 0.23, 0.09, 0.08, 0.05 | |
| 10 | 14 | 15 | 2.01, 1.81, 1.72, 1.85, 1.59 * | 0.11, 0.06, 0.07, 0.10, 0.09 | |
| 20 | 14 | 15 | 1.08, 1.37, 1.04, 0.34, 1.06 * | 0.15, 0.10, 0.10, 0.14, 0.17 | |
| QUIN, short-term | 0 | 3 | 4 | 5.45, 5.58, 4.52, 4.81, 4.53, 5.84 | 0.06, 0.12, 0.07, 0.18, 0.10, 0.10 |
| 75 | 3 | 4 | 2.35, 3.18, 3.31, 3.94, 3.05, 1.9 * | 0.06, 0.10, 0.03, 0.10, 0.04 | |
| 100 | 3 | 4 | 1.55, 1.76, 2.12, 4.49, 4.11, 3.38 * | 0.07, 0.09, 0.09, 0.11, 0.10, 0.06 | |
| 125 | 3 | 4 | 2.82, 1.42, 4.25, 4.64, 1.44, 3.45 * | 0.06, 0.11, 0.09, 0.09, 0.07, 0.05 | |
| QUIN, protracted | 0 | 14 | 15 | 2.79, 3.36, 2.36, 2.18, 2.18, 2.53 | 0.08, 0.10, 0.07, 0.05, 0.07, 0.12 |
| 50 | 14 | 15 | 2.57, 2.29, 2.36, 2.13, 3.11, 2.34 | 0.09, 0.15, 0.12, 0.07, 0.12, 0.09 | |
| 75 | 14 | 15 | 2.83, 3.23, 2.20, 2.79, 1.67, 2.96 | 0.06, 0.11, 0.07, 0.09, 0.13, 0.18 | |
| 100 | 14 | 15 | 3.15, 2.91, 3.14, 3.50, 3.09, 3.08 | 0.11, 0.09, 0.09, 0.07, 0.11, 0.15 |
Abbreviations: DEN = diethylnitroamine; QUIN = quinoline; %RET = percent reticulocytes; %MN-RET = percent micronucleated reticulocytes.
= Treatment group is significantly different from concurrent vehicle control, p < 0.05, two-sided Dunnett’s test in the case of %RET, one-sided in the case of %MN-RET.
DEN: 14-day study
The 10 and 20 mg/kg/day DEN dose groups exhibited significant reductions to body weight gains as well as normalized liver weights (Figure 4a,b). As no other outward signs of toxicity were evident, it appears that the doses used in this 14-day study were well tolerated.
Figure 4.
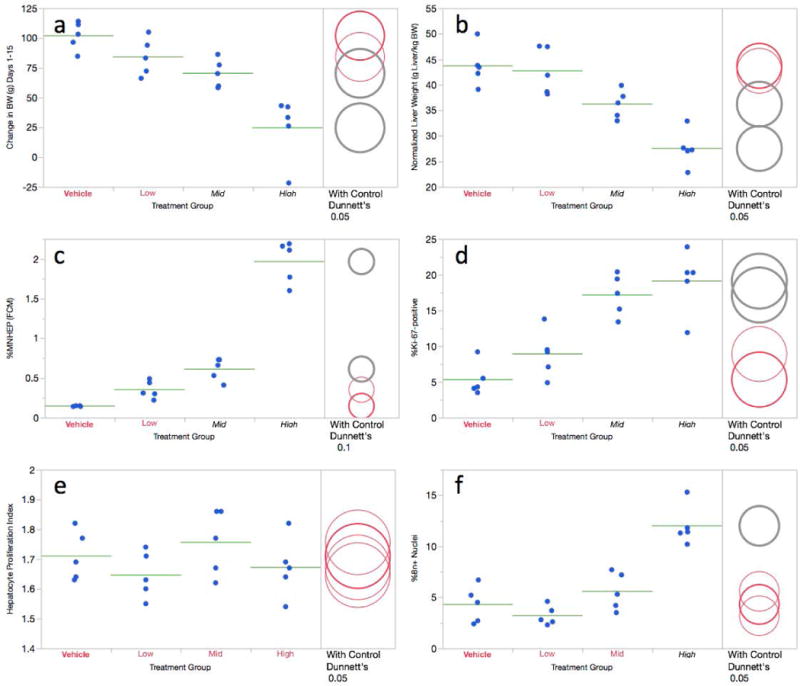
a-f Flow cytometric data for the 14 day diethylnitrosamine study are shown. Panel a = Change in body weight over the course of the treatment period; panel b = normalized liver weights; panel c = %MNHEP; panel d = %Ki-67-positive nuclei; panel e = hepatocyte proliferation index; panel f = %8n+ nuclei. For this study, low, mid, and high treatment groups correspond to 5, 10, and 20 mg/kg/day, respectively. For every graph, data for each individual rat is shown, and group means appear as horizontal green lines. Dunnett’s test results are shown to the far right of each graph, where statistically significant differences relative to the concurrent vehicle control group appear as italicized black text as opposed to red text, and by grey circles as opposed to red circles. Circles’ diameters represent 95% confidence intervals.
As shown by Figure 4c, flow cytometry-based %MNHEP values were observed to increase in a dose-dependent manner, with mid and high dose groups reaching statistical significance. Whereas the vehicle control animals averaged 0.15% MNHEP, rats in the 20 mg/kg/day dose group exhibited a mean frequency of 1.97%, or a 13.1-fold average increase. The highest dose level shared across the two DEN studies, on a per day basis, is 20 mg/kg. As compared to the 14-day study’s 13.1-fold increase, the short-term treatment design induced a 2.9-fold increase at this dose.
Regarding the recent hepatocyte division metric, Ki-67-positive nuclei frequency, the mid and high dose groups showed significant increases (Figure 4d). Whereas DEN treatment did not affect HPI (Figure 4e), 8n+ nuclei frequencies were significantly elevated in the high dose group (Figure 4f).
As was the case for the 3-day study, blood samples collected one day following cessation of 14-days of DEN treatment were evaluated for micronucleus induction. DEN exposure was found to significantly decrease %RET, whereas no effect was observed for MN-RET frequencies (Table I).
QUIN: 3-day study
Each of the three groups exposed to QUIN showed significant reductions to body weight gain (Figure 5a). Unlike DEN treatment that reduced normalized liver weight, the high dose QUIN group exhibited a marked increase (Figure 5b). No other outward signs of toxicity were evident, and we conclude that the dose levels chosen for this study were well tolerated.
Figure 5.
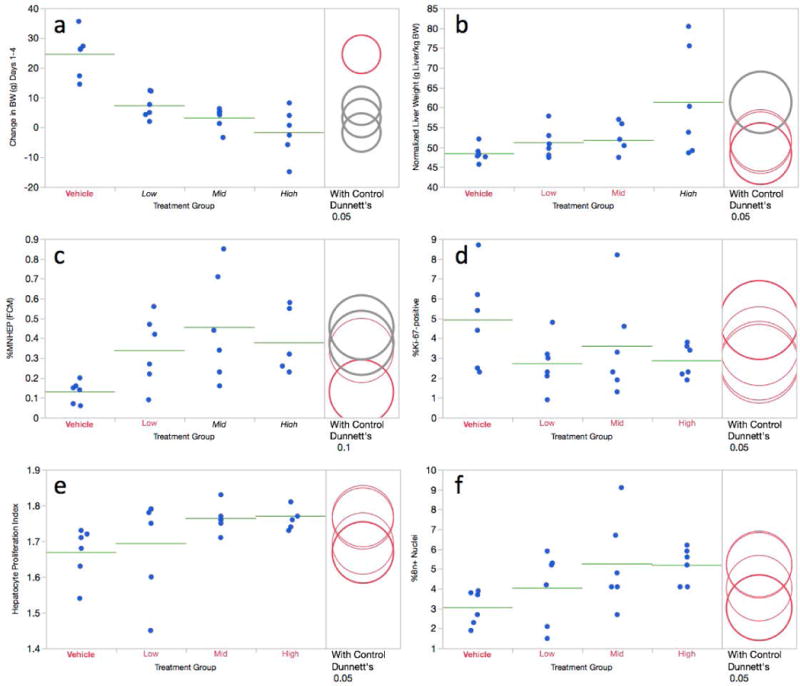
a-f. Flow cytometric data for the 3 day quinoline study are shown. Panel a = Change in body weight over the course of the treatment period; panel b = normalized liver weights; panel c = %MNHEP; panel d = %Ki-67-positive nuclei; panel e = hepatocyte proliferation index; panel f = %8n+ nuclei. For this study, low, mid, and high treatment groups correspond to 75, 100, and 125 mg/kg/day, respectively. For every graph, data for each individual rat is shown, and group means appear as horizontal green lines. Dunnett’s test results are shown to the far right of each graph, where statistically significant differences relative to the concurrent vehicle control group appear as italicized black text as opposed to red text, and by grey circles as opposed to red circles. Circles’ diameters represent 95% confidence intervals.
As shown by Figure 5c, flow cytometry-based %MNHEP values were elevated in the mid and high dose groups relative to vehicle control. The highest mean value, 0.46% MNHEP, was observed in the mid dose group (100 mg/kg/day), although considerable intra-group variation was evident.
The 3-day QUIN exposures did not cause statistically significant changes to hepatocyte proliferation metrics (Figure 5d,e,f). The absence of remarkable changes to these biomarkers, in light of increases to normalized liver weight, is suggestive of hypertrophy. This interpretation is supported by histological observations reported by Uno and colleagues in the Japanese JEMS/MMS Group’s QUIN study [Uno et al., 2015].
Blood samples collected one day following cessation of 3-days of QUIN treatment were evaluated for micronuclei. While QUIN exposure was found to significantly decrease %RET, no effect was observed for the MN-RET endpoint (Table I).
QUIN: 14-day study
The protracted treatment design resulted in significant reductions to body weight gains in rats exposed to the two highest dose levels (Figure 6a). All three of the QUIN-exposed groups showed marked increases in normalized liver weights (Figure 6b). As no additional signs of toxicity were evident, it appears that the doses used for this study were well tolerated.
Figure 6.

a-f. Flow cytometric data for the 14 day quinoline study are shown. Panel a = Change in body weight over the course of the treatment period; panel b = normalized liver weights; panel c = %MNHEP; panel d = %Ki-67-positive nuclei; panel e = hepatocyte proliferation index; panel f = %8n+ nuclei. For this study, low, mid, and high treatment groups correspond to 50, 75, and 100 mg/kg/day, respectively. For every graph, data for each individual rat is shown, and group means appear as horizontal green lines. Dunnett’s test results are shown to the far right of each graph, where statistically significant differences relative to the concurrent vehicle control group appear as italicized black text as opposed to red text, and by grey circles as opposed to red circles. Circles’ diameters represent 95% confidence intervals.
Flow cytometry-based MNHEP frequencies were elevated in the mid and high dose groups Figure 6c). The highest mean value, 0.62% MNHEP, was observed in both the mid and high dose QUIN groups (75 and 100 mg/kg/day, respectively), a value that corresponds to a 5.6-fold increase compared to vehicle control. The highest dose level shared across the two QUIN studies, on a per day basis, is 100 mg/kg. As compared to the 14-day study’s 5.6-fold increase, the short-term treatment design induced a 3.5-fold increase at this dose.
The three hepatocyte proliferation metrics, Ki-67-positive nuclei, HPI, and %8n+ nuclei, all showed substantial intra-group variation in treated animals (6d,e,f). Based on the variation and relatively modest differences compared to control rats, only %8n+ nuclei values in the high dose group attained statistical significance. As with the short-term study, lack of marked treatment-related effects on hepatocyte proliferation in this 14-day study, coupled with clear increases to normalized liver weight, suggests a QUIN-induced hypertrophy effect.
In addition to the liver-centric analyses described above, peripheral blood samples were analyzed for RET and MN-RET frequencies. Protracted QUIN exposure did not significantly affect either of these endpoints at the 15-day time point (Table I).
Flow cytometry versus microscopy
MNHEP frequencies generated by flow cyometry were directly compared to parallel measurements accomplished via acridine orange staining and microscopic inspection. Figure 7a graphically depicts the relationship that exists between these scoring methods over a wide range of %MNHEP frequencies. Whereas the flow cytometry-based measurements tended to produce slightly lower %MNHEP values compared to microscopy, the 0.80 r2 value provides strong evidence that the resulting data are comparable. While the frequencies are clearly in agreement, three differences are noteworthy. First, the measurements are collected at vastly different efficiencies. Whereas each microscopy-based analysis required an operator to spend approximately 20 minutes to interrogate approximately 2,000 hepatocytes per rat for the presence of micronuclei, the flow cytometry-based values were completed in approximately 2 minutes, and 20,000 nuclei were interrogated per sample. Considering both factors, this represents a 100-fold increase in data acquisition efficiency. Second, as described above for the DEN and QUIN studies, the flow cytometric method simultaneously provided several concurrent assessments of cell proliferation, an important aspect of any micronucleus assay. Third, by scoring the incidence of micronuclei in many times more cells, the distribution of baseline values generated by flow cytometry differed in significant ways. As shown by Figure 7b,c, the %MNHEP distribution was tighter for flow cytometry compared to microscopy. Specifically, the mean %MNHEP ± standard deviation (%coefficient of variation) was 0.14% ± 0.05 (36%) for the automated method, versus 0.05% ± 0.06 (120%) for microscopy. Furthermore, there were no zero %MNHEP readings for flow cytometry, compared to 14 out of 27, or slightly over 50% for manual scoring. Generally speaking, generating a high percentage of zero reading is undesirable, as it complicates the interpretation of modest changes over concurrent vehicle controls, and can negatively affect assays’ statistical power [OECD, 2014].
Figure 7.
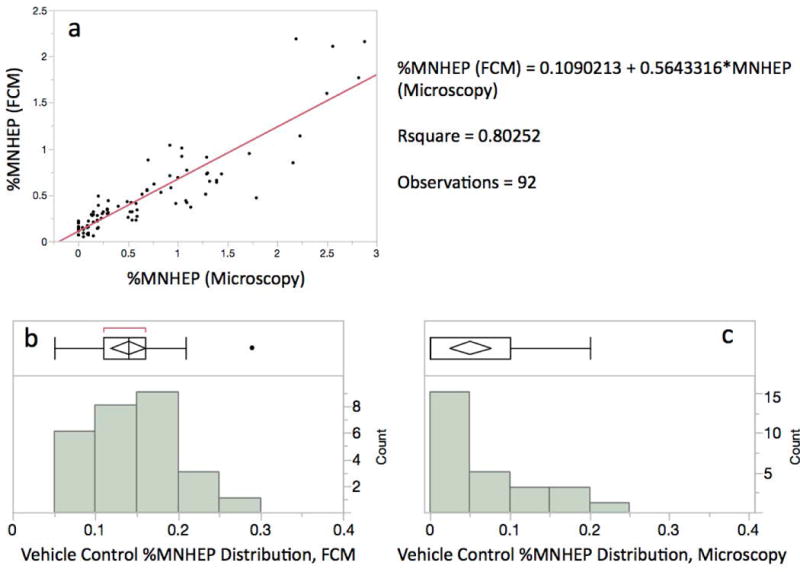
a-c. Panel a: Flow cytometry- and microscopy-based %MNHEP data are shown for split samples resulting from the two DEN and two QUIN studies, n = 92. The high r2 value suggests the methods produce highly correlated data. Panels b and c: %MNHEP frequency distribution plots for vehicle control animals based on flow cytometry and microscopy, respectively. Coefficient of variation and the number of readings without a zero %MNHEP result are improved with automated scoring.
Conclusions
While DEN and QUIN did not induce micronuclei in hematopoietic cells, they were observed to induce MNHEP in two different study designs, and similar results were generated by conventional microscopy and a novel flow cytometric method. This tissue specificity agrees with reports by the JEMS/MMS group [Uno et al., 2015a,b; Hamada et al., 2015], and the current data also supports their conclusion that there are some advantages to using a protracted dosing schedule when studying rats that are 6 weeks of age at time of treatment. That being said, clear induction of MNHEP using an acute treatment schedule were observed, and short-term experimental designs may have utility when training laboratory personnel on aspects of MNHEP enumeration, as well as for hazard identification, so long as animals’ age is chosen carefully with respect to the proportion of dividing cells.
Regarding the point about cell division, above, it is important to note that beyond generating reliable %MNHEP data, the multi-parametric nature of flow cytometry provided opportunities to collect useful data surrounding hepatocyte proliferation. More research is necessary, but it appears that the assay will benefit from Ki-67, HPI, and/or 8n+ nuclei readings, as they address division history, an essential consideration of any micronucleus assay.
Further work will be necessary to expand the number of chemicals studied using this automated scoring technique. In terms of genotoxic agents, aneugens as well as additional, diverse clastogens should be studied in order to better delineate the types of chromosome damaging-agents the assay is capable of detecting. Regarding assessments of assay specificity, presumed non-genotoxicants that cause various types of liver injury need to be studied. It will also be important to consider different treatment/harvest schedules, especially those that facilitate combining multiple genotoxicity endpoints into one acute study, or else integrating MNHEP assessment into ongoing repeat-dose toxicology studies, as these two strategies greatly reduce resource requirements and number of animals tested [Pfuhler et al., 2009; Hagio et al., 2014; Hamada et al., 2015]. The availability of an automated scoring method provides greater efficiencies for all such investigations, especially in terms of high quality, objective data acquisition. Finally, to achieve the greatest possible impact, inter-laboratory experiments will be necessary to evaluate the transferability of the automated scoring method described herein.
Supplementary Material
Acknowledgments
This work was funded by a grant from the National Institute of Health/National Institute of Environmental Health Sciences (NIEHS; grant no. R44ES026464). The contents are solely the responsibility of the authors, and do not necessarily represent the official views of the NIEHS. Several colleagues provided important intellectual support for which we are grateful, especially Marie Vasquez, Makoto Hayashi, Shuichi Hamada, and Rie Takashima.
Footnotes
Author contributions
S.D.D. and J.C.B. primarily designed these studies. S.L.A., S.K., P.S., and D.K.T. executed various aspects of the experiments. S.D.D. primarily wrote the manuscript, with significant input from all authors.
Disclosure
The authors are employees of Litron Laboratories. Litron has filed a patent covering flow cytometric methods for scoring micronucleated hepatocytes and plans to sell kits based on this technology (In Vivo MicroFlow® PLUS-RL Kits).
References
- Asano N, Katsuma Y, Tamura H, Higashikuni N, Hayashi M. An automated new technique for scoring the rodent micronucleus assay: computerized image analysis of acridine orange supravitally stained peripheral blood cells. Mutat Res. 1998;404:149–154. doi: 10.1016/s0027-5107(98)00108-0. [DOI] [PubMed] [Google Scholar]
- Chang Y, Zhou C, Huang F, Torous DK, Luan Y, Shi C, Wang H, Wang X, Wei N, Xia Z, Zhong Z, Zhang M, An F, Cao Y, Geng X, Jiang Y, Ju Q, Yu Y, Zhu J, Dertinger SD, Li B, Liao M, Yuan B, Zhang T, Yu J, Zhang Z, Wang Q, Ma J. Inter-laboratory validation of the in-vivo flow cytometric micronucleus analysis method (MicroFlow®) in China. Mutat Res. 2014;772:6–13. doi: 10.1016/j.mrgentox.2014.04.027. [DOI] [PubMed] [Google Scholar]
- Dertinger SD, Camphausen K, MacGregor JT, Bishop ME, Torous DK, Avlasevich S, Cairns S, Tometsko CR, Menard C, Muanza T, Chen Y, Miller RK, Cederbrant K, Sandelin K, Pontén I, Bolcsfoldi G. Three-color labeling method for flow cytometric measurement of cytogenetic damage in rodent and human blood. Environ Mol Mutagen. 2004;44:427–35. doi: 10.1002/em.20075. [DOI] [PubMed] [Google Scholar]
- Dertinger SD, Bishop ME, McNamee JP, Hayashi M, Suzuki T, Asano N, Nakajima M, Saito J, Moore M, Torous DK, MacGregor JT. Flow cytometric analysis of micronuclei in peripheral blood reticulocytes: I. Intra- and interlaboratory comparison with microscopic scoring. Toxicol Sci. 2006;94:83–91. doi: 10.1093/toxsci/kfl075. [DOI] [PubMed] [Google Scholar]
- Grawé J, Zetterberg G, Amnéus H. Flow-cytometric enumeration of micronucleated polychromatic erythrocytes in mouse peripheral blood. Cytometry. 1992;13:750–758. doi: 10.1002/cyto.990130711. [DOI] [PubMed] [Google Scholar]
- Hagio S, Furukawa S, Abe M, Kuroda Y, Hayashi S, Ogawa I. Repeat dose liver micronucleus assay using adult mice with multiple genotoxicity assays concurrently performed as a combination test. J Toxicol Sci. 2014;39:437–445. doi: 10.2131/jts.39.437. [DOI] [PubMed] [Google Scholar]
- Hamada S, Ohyama W, Takashima R, Shimada K, Matsumoto K, Kawakami S, Uno F, Sui H, Shimada Y, Imamura T, Matsumura S, Sanada H, Inoue K, Muto S, Ogawa I, Hayashi A, Takayanagi T, Ogiwara Y, Maeda A, Okada E, Terashima Y, Takasawa H, Narumi KY, Wako K, Kawasako M, Sano Ohashi N, Morita T, Kojima H, Honma M, Hayashi M. Evaluation of the repeated-dose liver and gastrointestinal tract micronucleus assays with 22 chemicals using young adult rats: Summary of the collaborative study by the Collaborative Study Group for the Micronucleus Test (CSGMT)/The Japanese Environmental Mutagen Society (JEMS) Mammalian Mutagenicity Study Group (MMS) Mutat Res. 2015;780-781:2–17. doi: 10.1016/j.mrgentox.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Hayashi M, MacGregor JT, Gatehouse DG, Blakey DH, Dertinger SD, Abramsson-Zetterberg L, Krishna G, Morita T, Russo A, Asano N, Suzuki H, Ohyama W, Gibson D. In vivo erythrocyte micronucleus assay: III. Validation and regulatory acceptance of automated scoring and the use of rat peripheral blood reticulocytes, with discussion of non-hematopoietic target cells and a single dose-level limit test. Mutat Res. 2007;627:10–30. doi: 10.1016/j.mrgentox.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Heddle J. A rapid in vivo test for chromosome damage. Mutat Res. 1973;18:187–190. doi: 10.1016/0027-5107(73)90035-3. [DOI] [PubMed] [Google Scholar]
- Higami Y, Shimokawa I, Okimoto T, Tomita M, You T, Ikeda T. Effect of aging and dietary restriction on hepatocyte proliferation and death in male F344 rats. Cell and Tissue Research. 1997;288:69–77. doi: 10.1007/s004410050793. [DOI] [PubMed] [Google Scholar]
- ICH. S2(R1) Guidance on genotoxicity testing and data interpretation for pharmaceuticals intended for human use. International Conference on Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use. ICH Harmonised Tripartite Guideline. 2011 Nov 9; Step 4. 2011. [Google Scholar]
- Kang JS, Wanibuchi H, Morimura K, Gonzalez FJ, Fukushima S. Role of CYP2E1 in diethylnitrosamine-induced hepatocarcinogenesis in vivo. Cancer Res. 2007;67:11141–11146. doi: 10.1158/0008-5472.CAN-07-1369. [DOI] [PubMed] [Google Scholar]
- Kirsch-Volders M, Sofuni T, Aardema M, Albertini S, Eastmond D, Fenech M, Ishidate M, Jr, Kirchner S, Lorge E, Morita T, Norppa H, Surralles J, Vanhauwaert A, Wakata A. Report from the in vitro micronucleus assay working group. Mutat Res. 2003;540:153–63. doi: 10.1016/j.mrgentox.2003.07.005. [DOI] [PubMed] [Google Scholar]
- MacGregor JT, Wehr CM, Gould DH. Clastogen-induced micronuclei in peripheral blood erythrocytes: The basis of an improved micronucleus test. Environ Mol Mutagen. 1980;2:509–514. doi: 10.1002/em.2860020408. [DOI] [PubMed] [Google Scholar]
- Matter B, Schmid W. Trenimon-induced chromosomal damage in bone marrow cells of six mammalian species, evaluated by the micronucleus test. Mutat Res. 1971;12:417–425. doi: 10.1016/0027-5107(71)90092-3. [DOI] [PubMed] [Google Scholar]
- OECD. Statistical analysis supporting the revisions of the genotoxicity Test Guidelines. OECD Publishing; Paris: 2014. (OECD Environment, Health and Safety Publications (EHS), Series on Testing and Assessment, No. 198). [Google Scholar]
- Pfuhler S, Kirkland D, Kasper P, Hayashi M, Vanparys P, Carmichael P, Dertinger S, Eastmond D, Elhajouji A, Krul C, Rothfuss A, Schoening G, Smith A, Speit G, Thomas C, van Benthem J, Corvi R. Reduction of use of animals in regulatory genotoxicity testing: Identification and implementation opportunities-Report from an ECVAM workshop. Mutat Res. 2009;680:31–42. doi: 10.1016/j.mrgentox.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Uno Y, Morita T, Luijten M, Beevers C, Hamada S, Itoh S, Ohyama W, Takasawa H. Recommended protocols for the liver micronucleus test: Report of the IWGT working group. Mutat Res. 2015a;783:13–18. doi: 10.1016/j.mrgentox.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Uno F, Tanaka J, Ueda M, Nagai M, Fukumuro M, Natsume M, Oba M, Akahori A, Masumori S, Takami S, Wako Y, Kawasako K, Kougo Y, Ohyama W, Narumi K, Fujiishi Y, Okada E, Hayashi M. Repeat-dose liver and gastrointestinal tract micronucleus assays for quinoline in rats. Mutat Res. 2015b;780-781:51–55. doi: 10.1016/j.mrgentox.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Reigh G, McMahon H, Ishizaki M, Ohara T, Shimane K, Esumi Y, Green C, Tyson C, Ninomiya S. Cytochrome P450 species involved in the metabolism of quinoline. Carcinogenesis. 1996;17:1989–1996. doi: 10.1093/carcin/17.9.1989. [DOI] [PubMed] [Google Scholar]
- Styles JA, Clark H, Festing MFW, Rew DA. Automation of mouse micronucleus genotoxicity assay by laser scanning cytometry. Cytometry. 2001;44:153–155. doi: 10.1002/1097-0320(20010601)44:2<153::aid-cyto1095>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Tometsko AM, Torous DK, Dertinger SD. Analysis of micronucleated cells by flow cytometry. 1. Achieving high resolution with a malaria model. Mutat Res. 1993;292:129–135. doi: 10.1016/0165-1161(93)90140-u. [DOI] [PubMed] [Google Scholar]
- Torous DK, Hall NE, Murante FG, Gleason SE, Tometsko CR, Dertinger SD. Comparative scoring of micronucleated reticulocytes in rat peripheral blood by flow cytometry and microscopy. Toxicol Sci. 2003;74:309–314. doi: 10.1093/toxsci/kfg143. [DOI] [PubMed] [Google Scholar]
- Torous DK, Hall NE, Illi-Love AH, Diehl MS, Cederbrant K, Sandelin K, Pontén I, Bolcsfoldi G, Ferguson LR, Pearson A, Majeska JB, Tarca JP, Hynes GM, Lynch AM, McNamee JP, Bellier PV, Parenteau M, Blakey D, Bayley J, van der Leede BJ, Vanparys P, Harbach PR, Zhao S, Filipunas AL, Johnson CW, Tometsko CR, Dertinger SD. Interlaboratory validation of a CD71-based flow cytometric method (MicroFlow) for the scoring of micronucleated reticulocytes in mouse peripheral blood. Environ Mol Mutagen. 2005;45:44–55. doi: 10.1002/em.20081. [DOI] [PubMed] [Google Scholar]
- Witt KL, Livanos E, Kissling GE, Torous DK, Caspary W, Tice R, Recio L. Comparison of flow cytometry- and microscopy-based methods for measuring micronucleated reticulocyte frequencies in rodents treated with nongenotoxic and genotoxic chemicals. Mutat Res. 2008;649:101–113. doi: 10.1016/j.mrgentox.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


