Fig. 5.
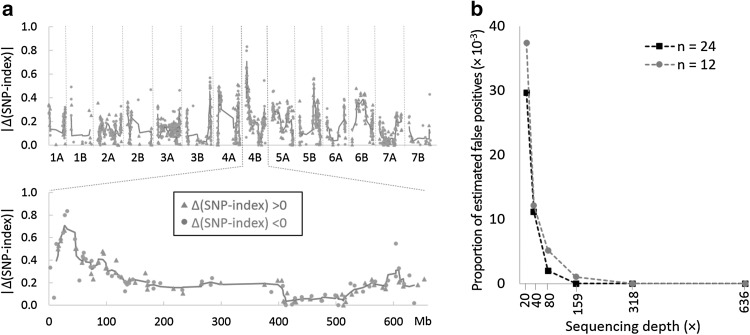
BSA and subsampling simulations under different sequencing depths and bulk sizes. a |Δ(SNP-index)| at each SNP position across the 14 tetraploid wheat chromosomes, with a detailed plot of chromosome 4B (sequencing depth = 1320×, bulk size = 24). Δ(SNP-index) at each position is calculated by subtracting the SNP-index (mutant allele frequency) in the W-bulk (24 shortest M4 lines) from that in the M-bulk (24 tallest M4 lines) [= SNP-index (M-bulk) − SNP-index (W-bulk)]. Blue triangles indicate SNPs with higher frequencies in the mutant bulk [SNP-index (M-bulk) > SNP-index (W-bulk)], while orange circles indicate SNPs with higher frequencies in the W-bulk [SNP-index (W-bulk) > SNP-index (M-bulk)]. Sliding window average (red) is plotted by averaging the |Δ(SNP-index)| values of five consecutive SNPs and shifting the window by one SNP at a time. b Proportion of false positives estimated by counting the number of SNPs with |Δ(SNP-index)| ≥0.8 outside the peak candidate region (chromosome 4B, 12.5–95.3 Mb; Table 1, Online Resource Text S1). (Color figure online)