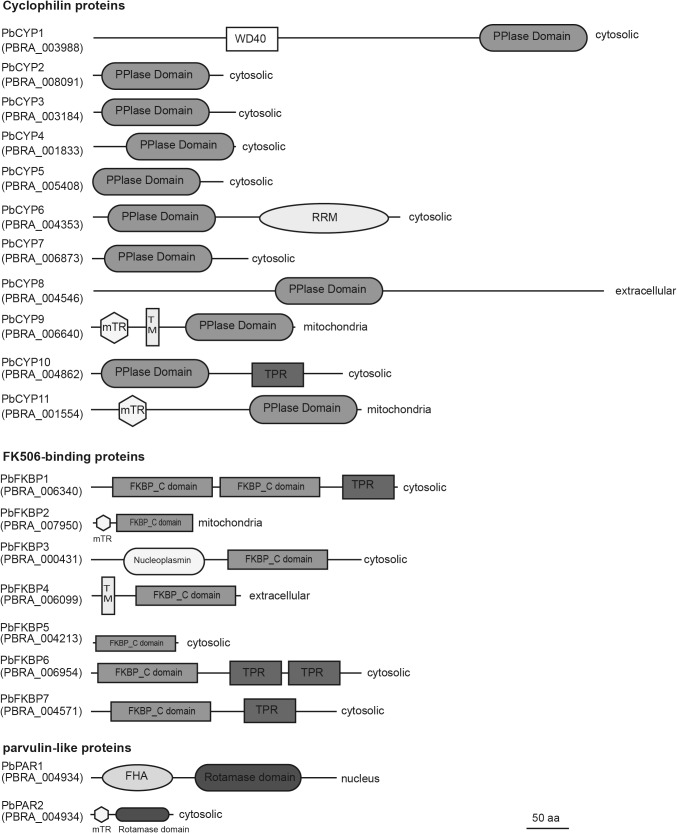
Fig. 1.
Domain architecture of predicted immunophilins in Plasmodiophora brassicae genome. Amino acid sequences were analyzed for the presence of conserved domains using the SMART software. WD40 tryptophan-aspartic acid repeats, PPIase peptidyl-prolyl isomerase, RRM RNA recognition motif, FKBP FK506-binding protein, TPR tetracopeptide repeat, FHA forkhead-associated domain, rotamase domain parvulin-like rotamase domain, mTP mitochondrial target peptide, TM transmembrane domain. Predicted cell localization is indicated. The bar marker indicates a length of 50 amino acids and refers to total protein length