Summary
Obligate intracellular parasites must efficiently invade host cells in order to mature and be transmitted. For the malaria parasite Plasmodium falciparum, invasion of host red blood cells (RBCs) is essential. Here we describe a parasite-specific transcription factor PfAP2-I, belonging to the Apicomplexan AP2 (ApiAP2) family, that is responsible for regulating the expression of genes involved in RBC invasion. Our genome-wide analysis by ChIP-seq shows that PfAP2-I interacts with a specific DNA motif in the promoters of target genes. Although PfAP2-I contains three AP2 DNA-binding domains, only one is required for binding of the target genes during blood stage development. Furthermore, we find that PfAP2-I associates with several chromatin-associated proteins, including the Plasmodium bromodomain protein PfBDP1, and that complex formation is associated with transcriptional regulation. As a key regulator of red blood cell invasion, PfAP2-I represents a potential new antimalarial therapeutic target.
Graphical abstract
Invasion of red blood cells is a highly regulated and essential process in the lifecycle of the malaria parasite Plasmodium falciparum. Santos et al. identify a transcription factor (PfAP2-I) that regulates invasion genes during blood stage development and associates with P. falciparum Bromodomain Protein 1 (PfBDP1).
Introduction
The phylum Apicomplexa consists of obligate intracellular eukaryotic parasites, including Plasmodium falciparum, the major causative agent of malaria. In the human host, P. falciparum merozoites invade RBCs, in which they grow and divide by schizogony during the 48-hour intraerythrocytic developmental cycle (IDC). Following replication, schizonts rupture and release 16–32 newly formed merozoites (Reilly et al., 2007), which egress from the RBC prepackaged with the proteins necessary for a new round of RBC invasion. Malaria symptoms, including fever and anemia, are correlated with the periodic and exponentially increasing waves of merozoite egress and invasion.
RBC invasion by the malaria parasite involves an orchestrated sequence of protein-protein interactions between the parasite and its host. Proteins mediating invasion decorate the merozoite surface or are stored at the apical end of the parasite in specialized secretory organelles named micronemes and rhoptries (figure 1A) (Gao et al., 2013; Singh and Chitnis, 2012). Of the ten known merozoite surface proteins (PfMSP1-10), PfMSP1 is the most abundant, with roles in merozoite egress (Das et al., 2015), budding (Combe et al., 2009) and likely the first steps of RBC attachment (Weiss et al., 2015). Stable adhesion of the parasite to RBCs is further mediated by proteins secreted from the rhoptry necks (the reticulocyte homologues, or Rhs), and proteins originating in the micronemes (the erythrocyte-binding like proteins, or EBLs), through specific interactions with host cell receptors (Tham et al., 2012). The rhoptry neck proteins (RONs), on the other hand, form a complex with the micronemal protein apical membrane antigen (PfAMA1) at the tight junction formed at the site of parasite attachment to the RBC membrane (reviewed in (Proellocks et al., 2010)). Merozoite invasion is an active process propelled by the glideosome, which is anchored between the plasma membrane and the inner membrane complex (IMC). This is a large molecular machine composed of a myosin motor (PfMyoA), a myosin light chain (myosin A tail domain-interacting protein, or PfMTIP) and several glideosome-associated proteins (GAPs) (Frenal et al., 2010). The glideosome-associated membrane proteins (GAPMs) also localize to the IMC but are only loosely associated with the glideosome (Bullen et al., 2009; Harding et al., 2016). Duffy binding-like MSPs (DBLMSPs), the rhoptry-associated proteins (RAPs) and the high molecular weight rhoptry proteins (RhopHs) are also important for invasion but their precise functions are unknown (reviewed in (Cowman et al., 2012)) (figure 1A).
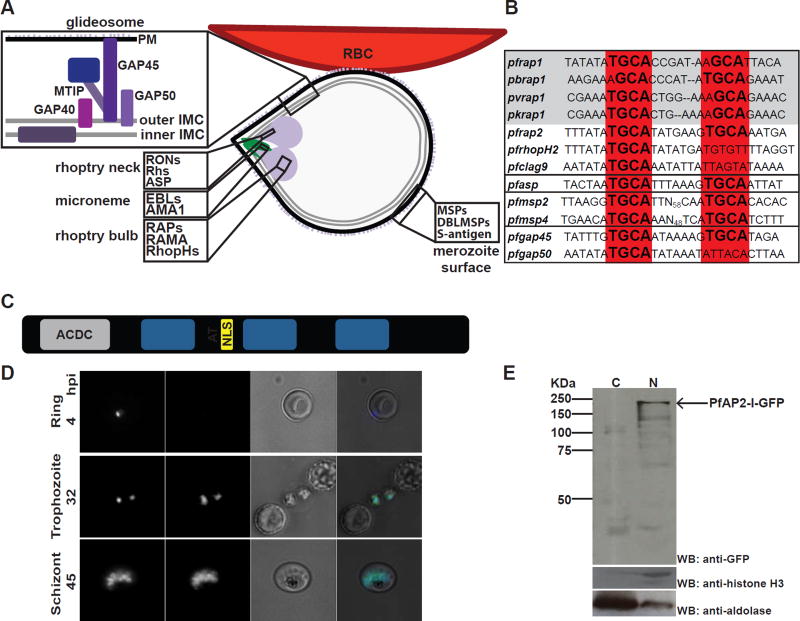
Figure 1. PfAP2-I is a nuclear protein that may bind a conserved TGCA DNA motif upstream of some invasion genes.
A- Merozoite attached to the RBC surface with the micronemes, rhoptries, merozoite surface, plasma membrane (PM), inner and outer inner membrane complex (IMC) depicted. Proteins found in the glideosome, merozoite surface, microneme, rhoptry neck and rhoptry bulb are highlighted. B- The NGGTGCA DNA sequence motif is conserved in invasion-related gene promoters across P. falciparum (Pf), P. berghei (Pb), P. vivax (Pv) or P. knowlesi (Pk) (grey box). The four black boxes (from top to bottom) contain rhoptry promoters, the pfasp rhoptry neck promoter, merozoite surface (msp) promoters, and glideosome (gap) promoters (adapted from (Young et al., 2008)) (see also figure S1A). C- The PfAP2-I protein structure contains a putative ACDC domain, the three AP2 DNA-binding domains, an AT-hook and a nuclear localization signal (NLS) (see also figure S2). D- Live fluorescence microscopy of synchronized parasites shows that PfAP2-I-GFP (see figure S1B) localizes to the nucleus of trophozoite and schizont stage parasites but is not detected in ring stages. Hoechst was used as a nuclear marker. BF denotes bright field. E- Nuclear fractionation followed by Western blot of schizont-stage PfAP2-I-GFP parasites confirms nuclear localization of PfAP2-I-GFP. Anti-histone H3 and anti-aldolase were used as nuclear and cytosolic markers, respectively. C: cytosol, N: nucleus.
Because invasion mechanisms are highly conserved among Plasmodium species (Weiss et al., 2015) and the merozoite is one of the few extracellular stages during the life cycle, proteins involved in invasion are attractive antimalarial targets. However, blocking RBC invasion is complex. The invasion process is fast and, with few notable exceptions, proteins that mediate parasite binding to the host RBC are functionally redundant (Walker et al., 2014). Merozoite surface proteins are also considered poor vaccine candidates because circulating merozoites express different surface variants, and the parasite can use alternative invasion pathways (reviewed in (Wright and Rayner, 2014)). Therefore, identifying conserved, essential invasion factors that can be therapeutically targeted remains a priority.
The expression of invasion-related genes is highly coordinated during daughter cell budding in Plasmodium parasites (Bozdech et al., 2003; Le Roch et al., 2003). Two conserved DNA elements are enriched upstream of invasion-related genes (Elemento et al., 2007; Essien and Stoeckert, 2010; Harris et al., 2011; Iengar and Joshi, 2009; Russell et al., 2015; Young et al., 2008). While the ACAACT motif (PfM20.1) is enriched in the promoters of micronemal genes, an NGGTGCA motif (PfM18.1) is present upstream of rhoptry genes (Young et al., 2008). The “rhoptry” motif is conserved among different Plasmodium species (figures 1B and S1A), and is present upstream of the msp gene family and other invasion-related genes but is not found in ron gene promoters (with the exception of apical sushi protein, or ASP), (figure 1B). This motif conservation implies that there are stage-specific transcription factors (TFs) that interact with these DNA motifs for expression of the invasion genes.
The ApiAP2 protein family is of plant origin and includes 27 sequence-specific DNA-binding proteins, expressed at all stages of the life cycle, each containing 1–3 AP2 DNA-binding domains (Balaji et al., 2005; Painter et al., 2011). In vitro studies have identified two AP2 domains from different ApiAP2 proteins that bind a GTGCA motif resembling the “rhoptry” motif (Campbell et al., 2010). The first of these proteins, PfSIP2 (PF3D7_0604100) associates uniquely with the SPE2 motifs found at the chromosome ends in the telomere-associated repetitive elements (TAREs) and upstream of var genes (Flueck et al., 2010). The second protein, PF3D7_1007700, is a 183 KDa protein, containing three AP2 domains, the third of which binds the “rhoptry” motif (Campbell et al., 2010). Here, we demonstrate that PF3D7_1007700 or PfAP2-I (ApiAP2 involved in invasion) is a key regulator of RBC invasion by the malaria parasite.
Results
PfAP2-I binds upstream of invasion-related genes
Homologues of the PfAP2-I protein are found across all Plasmodium species (Figure S2). PfAP2-I contains three conserved protein domains in addition to the three AP2 domains: an N-terminal ACDC domain (AP2-Coincident Domain mostly at the C-terminus) (Iwanaga et al., 2012; Oehring et al., 2012), a putative AT-hook DNA-binding domain between the first and second AP2 domains and a predicted nuclear localization signal (NLS) (figures 1C and S2). Knowing that domain 3 of PfAP2-I binds the NGGTGCA “rhoptry motif” in vitro (Campbell et al., 2010) we sought to determine if PfAP2-I binds this motif in vivo to regulate the transcription of invasion-related genes. First, we generated a parasite line expressing PfAP2-I C-terminally tagged with GFP (PfAP2-I-GFP) (figure S1B). Live microscopy and nuclear fractionation assays show that PfAP2-I localizes exclusively to the nucleus of trophozoite and schizont stage parasites (figures 1D-E).
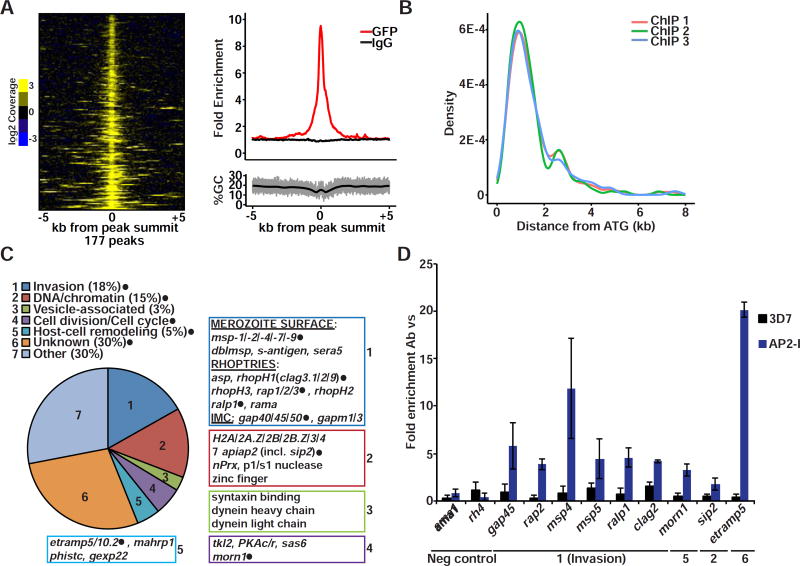
To determine the genome-wide binding sites for PfAP2-I, we performed three independent chromatin immunoprecipitation assays followed by high-throughput sequencing (ChIP-seq) from PfAP2-I-GFP expressing parasites with anti-GFP or non-immune IgG antibodies. Analysis of the ChIP-seq data identified 177 regions bound by PfAP2-I in at least two of three biological replicates (figure 2A and table S2). These 177 regions correspond to the upstream regions of 157 genes and are largely located 1–2 kb upstream of the ATG (figure 2B). While 30% of the PfAP2-I-bound genes encode proteins of unknown function, the largest functional group (18%) encodes known invasion proteins (figure 2C). PfAP2-I binds to the promoters of members of the msp, rap and rhopH gene families but it does not bind upstream of micronemal or ron genes (figure 2C and table S2). Gene ontology (GO) term analysis supported the enrichment of invasion-related genes and also revealed that nucleosome and chromatin-related gene promoters are targeted by PfAP2-I (table S2). Interestingly, PfAP2-I binds the promoters of seven apiap2 genes (figure 2C and table S2) including its own, suggesting that it may auto-regulate its own gene expression. Five of these apiap2 genes are transcribed immediately following pfap2-I during the IDC (Campbell et al., 2010). We also found PfAP2-I upstream of cell division/cell cycle-related genes encoding proteins important during schizogony and genes related to vesicle formation and host cell remodeling (figure 2C). We validated our ChIP-seq data by ChIP-qPCR and confirmed that PfAP2-I does not bind regions bound by PfSIP2 (figures 2D and S3A) (Flueck et al., 2010). This suggests that although PfAP2-I and PfSIP2 bind a similar DNA motif in vitro (Campbell et al., 2010), they target distinct genome-wide regions. These results demonstrate that PfAP2-I is associated with the promoters of a specific set of genes that are enriched in invasion-related functions.
Figure 2. PfAP2-I-GFP binds upstream of invasion genes by ChIP.
A- Heatmap (left) of 165 genomic loci from ChIP-seq replicate 1, +/− 5kb of each anti-GFP peak summit found in at least two of three replicates, shows enrichment of sequence reads (PfAP2-I bound) on ChIP/input samples (shown is the log2 transformed coverage). Profile plot from replicate 1 (top right) shows enrichment of anti-GFP ChIP-seq (red) compared to the control IgG ChIP-seq (black). The increase in signal is unrelated to GC content (bottom right). B- Plot showing the position of ChIP-seq peak summits relative to the ATG in the three replicates. C- Functional categorization of the 157 ChIP-Seq target genes based on literature review and GO term enrichment. Annotated examples of genes belonging to each category are shown (genes tested in D are highlighted with filled black circles). For the full list of gene targets and GO term results see Table S2. D- ChIP-qPCR of selected genes confirms PfAP2-I binding (blue bars) to ChIP-seq targets and no binding to the ama1 and rh4 promoters. Although not detected by ChIP-seq (see figure S3D), PfAP2-I associates with msp5 according to ChIP-qPCR. 3D7 wt parasites were used as a negative control (black bars). The results are shown as fold enrichment of ChIP performed with antibody versus non-immune IgG (n=3). Data are represented as mean ± SD (see also figure S3A).
DNA-binding by PfAP2-I is important for gene transcription and only requires one AP2 domain
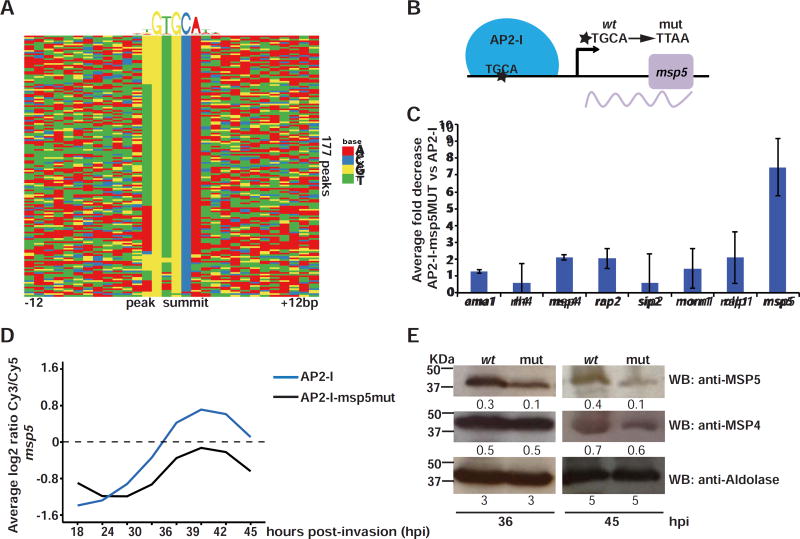
Analysis of the DNA motifs enriched in the PfAP2-I ChIP-seq target gene sequences identified six DNA motifs with GTGCA being the most significant (2E−25) (figures 3A and S3B). The GTGCA motif is highly similar to the motif bound in vitro by domain 3 of PfAP2-I (Campbell et al., 2010) (figure S3B). Four of the additional motifs are highly degenerate while a sixth, ATGCA (8E−3), is analogous to the predominant PfAP2-I motif (figure S3B). To provide a direct link between gene transcription and binding of PfAP2-I, we tested whether mutating the GTGCA motif in a target gene promoter decreases transcription of that gene. Since P. falciparum is haploid and we anticipated that mutation of the motif would abolish or greatly reduce expression, this required testing a non-essential gene to maintain parasite viability. For this, we chose pfmsp5 (pf3d7_0206900). This gene has previously been knocked out (Sanders et al., 2006), contains a putative PfAP2-I DNA binding motif (ATGCA) in the promoter and is adjacent to two PfAP2-I ChIP-seq targets (pfmsp2 and pfmsp4) (figure S3D). Although pfmsp5 was not identified as a ChIP-seq target gene, ChIP-qPCR showed that PfAP2-I is enriched in the promoter of pfmsp5 (figure 2D). We used CRISPR/Cas9 to mutate the ATGCA motif in its promoter, generating the PfAP2-I-GFP::msp5MUT strain (figures 3B and S3E). In this line, binding of PfAP2-I to the mutated pfmsp5 promoter was severely reduced, as determined by ChIP-qPCR (figures 3C and S3F). Levels of pfmsp5 transcript were also greatly reduced, as measured by DNA microarrays and RT-qPCR (figures 3D and S3G-I). Although global transcription was not affected in the mutant line, we did detect an unexpected decrease in the levels of some transcripts (Rnits p-value <5E−2) (figure 3H and table S3), which we hypothesize is due to compensatory regulatory mechanisms. MSP5 protein levels reflected the drop in msp5 transcription and were decreased three-fold (figure 3E). PfAP2-I binding to other promoters (figures 3C and S3F) and translation levels of the proximal invasion gene product MSP4 were not affected (figure 3E), suggesting that PfAP2-I modulates transcription through direct and specific interaction with the TGCA DNA motif found upstream of its target genes.
Figure 3. PfAP2-I binding to the TGCA DNA motif is important for transcription.
A- DNA motif heatmap of the trimmed ChIP-seq peaks +/−12bp surrounding the highest scoring motif (as discovered by DREME, figure S3B) shows that the GTGCA DNA motif is found within the majority of the peaks. Each row represents a peak summit and each column represents an individual nucleotide (See also figures S3B-C and S4). B- The mutations introduced to generate PfAP2-I-GFP::msp5MUT parasites (see also figures S3D-E). C- ChIP-qPCR showing that PfAP2-I binding to msp5 in PfAP2-I-GFP::msp5MUT parasites is decreased more than 7-fold versus PfAP2-I-GFP parasites. The data are represented as mean ± SD and n=3 (see figure S3F for original fold values). D- Microarray data using RNA extracted from PfAP2-I-GFP and PfAP2-I-GFP::msp5MUT parasites at eight time points beginning at 18hpi (see figure S3G) shows that when the TGCA motif is mutated upstream of msp5, levels of the msp5 transcript are reduced (see also figures S3H-I). The plot shows the average of two biological replicates (See Table S3 for complete microarray results). E- Western blot using specific antibodies against MSP5 show that the MSP5 protein level is decreased in PfAP2-I-GFP::msp5MUT (mut) versus PfAP2-I-GFP (wt) parasites but MSP4 and aldolase protein levels are unchanged at 36 and 45hpi. Densitometry values are shown below each blot.
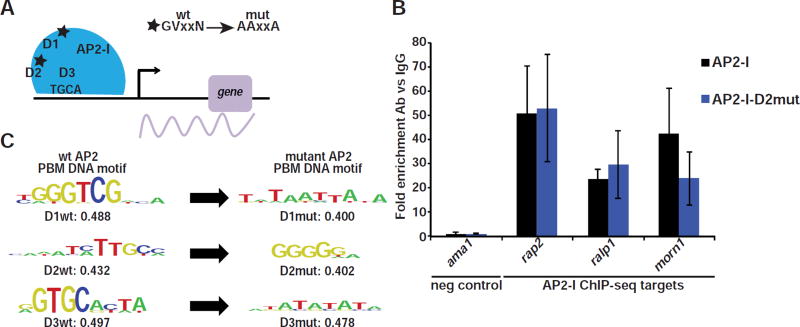
The full-length PfAP2-I protein contains three AP2 DNA-binding domains that bind distinct DNA motifs in vitro (figure 4C) (Campbell et al., 2010). Motif analysis of the 250bp sequences surrounding each predicted ChIP-seq peak summit revealed that the DNA motifs uniquely bound by the second (D2) and third domain (D3) of PfAP2-I are prevalent in many of the ChIP-seq peaks but that the motif bound by the first AP2 domain (D1) is less common (figures S4A-B). However, we failed to identify the D1 or D2 DNA motifs when performing de novo motif enrichment (figure S3B), leading us to hypothesize that D1 and D2 were not important for in vivo DNA-binding during the RBC stage. To test this, we attempted to disrupt the function of each of the three AP2 domains individually by generating parasite lines in which conserved amino acid residues were mutated to alanine using CRISPR/Cas9 mutagenesis (figures 4A and S5A). While we were able to obtain parasites carrying mutations in AP2-I D1 (PfAP2-I-GFP-D1mut) and in AP2-I D2 (PfAP2-I-GFP-D2mut), we repeatedly failed to generate PfAP2-I-GFP-D3mut parasites (figures S5B-D). Similarly, all attempts to disrupt pfap2-i (data not shown) or its orthologue in P. berghei (PBANKA_120590) (Modrzynska et al., 2017) were unsuccessful, suggesting that PfAP2-I is essential. By ChIP-qPCR we confirmed that in the PfAP2-I-GFP-D2mut parasites, PfAP2-I was functional and remained associated to the same set of target genes as in wt parasites (figure 4B). In parallel, we performed protein binding microarrays (PBMs) (Berger and Bulyk, 2009; Campbell et al., 2010) with GST-tagged AP2-D1, AP2-D2 and AP2-D3 proteins carrying the same mutations to ensure that the introduced mutations abolished DNA binding (figures 4C and S5C-D and Table S6). Therefore, during RBC development only the third AP2 domain of PfAP2-I, which binds GTGCA, is required to coordinate transcription.
Figure 4. Only PfAP2-I-D3 is required for DNA-binding during the IDC.
A- The mutations introduced in the AP2 domains resulting in PfAP2-I-GFP-D1mut and PfAP2-I-GFP-D2mut parasites (see also figures S5A-D). B- ChIP-qPCR with PfAP2-I-D2mut parasites shows that PfAP2-I still binds to its target genes when D2 is mutated (blue bars). PfAP2-I-GFP parasites were used as control (black bars). Data are represented as mean ± SD and n=3. C- PBMs show that the DNA-binding of PfAP2-I AP2 domains D1, D2 and D3 are altered when the domains are mutated. Shown are both wt and mutant PBM results. Enrichment values below 0.45 represent low affinity DNA-binding (see Table S6 for position weight matrices for all AP2 domains).
Gene transcription may require an interaction between PfAP2-I and PfBDP1
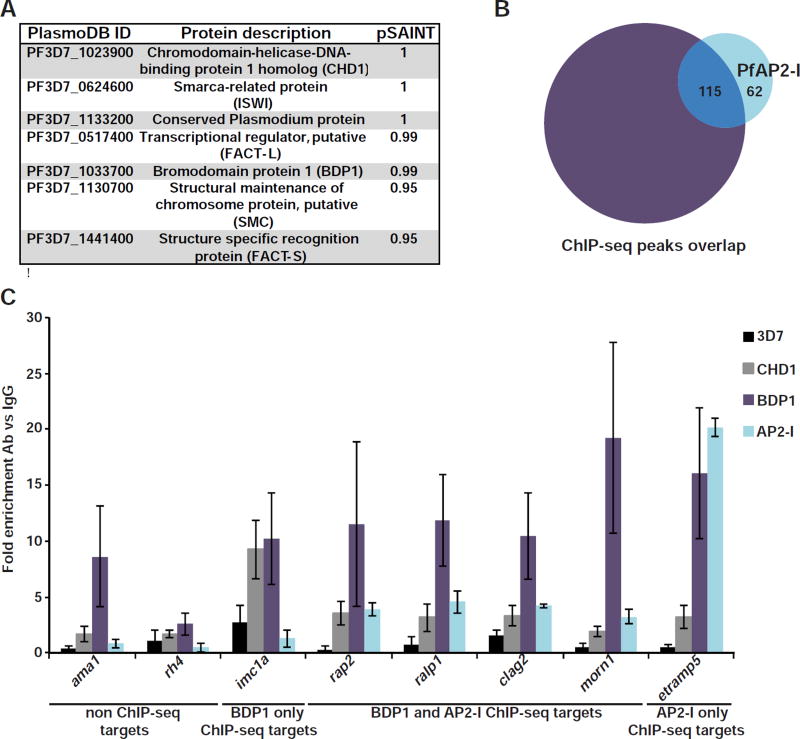
In order to recruit the transcriptional machinery, PfAP2-I likely interacts with other proteins in the cell. To determine its interaction partners, we performed immunoaffinity purification (IP) from PfAP2-I-GFP expressing parasites (Table S4 and Figure S5G) (Joshi et al., 2013). Among the high specificity protein interactions identified (pSAINT ≥0.9) (figure 5A and table S4), we found four that have been previously localized to the nucleus (Bischoff and Vaquero, 2010; Horrocks et al., 2009; Ji and Arnot, 1997; Josling et al., 2015; Templeton et al., 2004; Volz et al., 2010). These chromatin-associated proteins include Bromodomain Protein 1 (PfBDP1), which has been shown to positively regulate the transcription of invasion-related genes (Josling et al., 2015). Given our finding that PfBDP1 co-purified with PfAP2-I, we compared the repertoire of ChIP-seq peaks reported for PfBDP1-HA (Josling et al., 2015) to our PfAP2-I-GFP ChIP-seq results and found that 65% of PfAP2-I peaks overlap with PfBDP1 bound genomic regions (figure 5B and table S2). Combined with our IP data, this suggests that PfAP2-I and PfBDP1 may interact in a protein complex to coordinate gene expression.
Figure 5. PfAP2-I and PfBDP1 coimmunoprecipitate and regulate transcription of many of the same genes.
A- List of PfAP2-I interaction partners with probability (pSAINT) of interaction as measured by SAINT of 0.9 or above (see also figure S6A and for full list of proteins detected see Table S4). B- Venn diagram comparing the peaks found by ChIP-seq for PfBDP1-HA (Josling et al., 2015) (purple) and PfAP2-I-GFP (this study) (light blue) shows that 115 genomic regions are bound by both proteins. See also Table S2. C-ChIP-qPCR of PfAP2-I-GFP, PfBDP1-HA and PfCHD1-GFP confirms that PfAP2-I does not bind genes only found in the PfBDP1 ChIP-seq dataset. Data are represented as mean ± SD and n=4 (see also Figure S6B, which shows some of the same data).
Interestingly, PfBDP1 also binds many gene promoters not bound by PfAP2-I (figure 5B and table S2). Our re-analysis of the published PfBDP1 ChIP-seq data (Josling et al., 2015) found that PfBDP1 is enriched at a GTGCAH motif, as reported. However, a second DNA motif (CWTAACTW), which is similar to the “micronemal” DNA motif (ACAACCT) (Young et al., 2008) was also identified (figures S6B-D). These data suggest a model (figure 6) in which PfAP2-I and PfBDP1 form a complex and bind the GTGCAH DNA motif to regulate the expression of rhoptry and merozoite surface genes, but not micronemal genes. ChIP-qPCR with PfBDP1-HA parasites confirmed that PfBDP1 interacts with the 5’ intergenic region of invasion genes bound by PfAP2-I but also with other invasion-related genes not associated with PfAP2-I (figures 5C and S6A). Similarly, another protein detected by IP, Chromodomain Protein 1 (PfCHD1), binds additional promoters besides those bound by PfAP2-I, as determined by ChIP-qPCR using PfCHD1-GFP parasites (Volz et al., 2010) (figures 5C and S6A). Therefore, although PfAP2-I likely associates with PfBDP1 and PfCHD1, they may also be independently recruited to chromatin through other DNA-binding proteins.
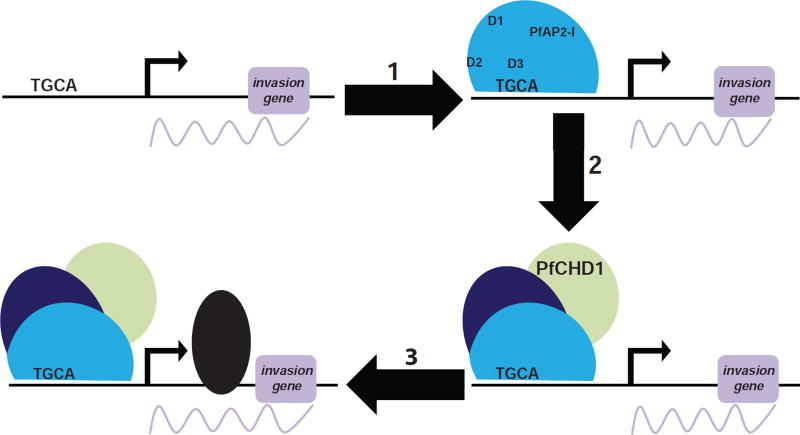
Figure 6. Model of transcription regulation by PfAP2-I and PfBDP1.
Our model is that PfAP2-I must first bind the TGCA DNA motif within the promoters of invasion genes via its third AP2 domain (D3). Subsequently, PfBDP1 and PfCHD1 are recruited followed by RNA polymerase II thereby initiating transcription.
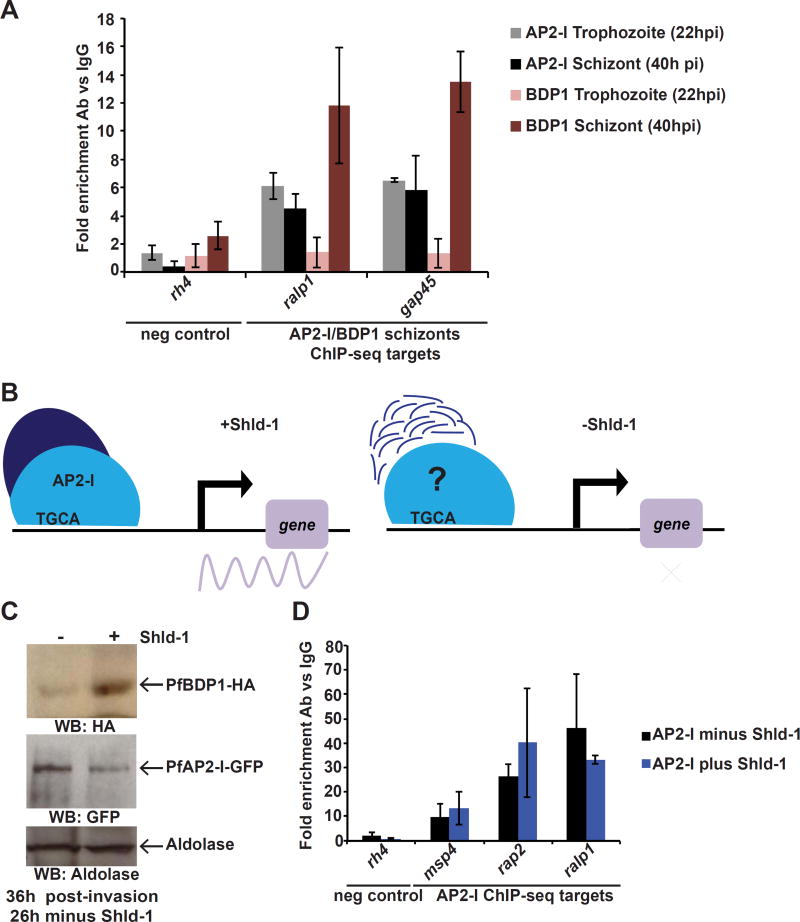
To determine the temporal association of the PfAP2-I/PfBDP1 complex on the chromatin, we explored whether PfAP2-I binds to the invasion promoters before or after PfBDP1. First, we tested if PfAP2-I and/or PfBDP1 were bound to the promoters at the trophozoite stage prior to transcription of the target genes. Indeed PfAP2-I binds to the target gene promoters at the trophozoite stage (22h post-invasion) and remains associated in schizonts (40h post-invasion). However, PfBDP1, as previously shown (Josling et al., 2015), does not bind at the trophozoite stage and only binds at the later time point (Figures 7A and S7A). We then pursued a knockdown strategy to test DNA-binding in the absence of one of the two proteins. While we were unable to generate a PfAP2-I knockdown (data not shown), PfBDP1 has been previously knocked down using the FK506 binding protein (FKBP) destabilization domain (DD) system (PfBDP1-HA-DD) (Josling et al., 2015). In these parasites, PfBDP1 is fused to DD, which is stabilized only in the presence of the small molecule ligand Shld-1 (Armstrong and Goldberg, 2007). PfBDP1-HA-DD parasites grown in the absence of Shld-1 invade RBCs poorly due to the reduced expression of invasion genes (Josling et al., 2015). To study the interaction between PfAP2-I and PfBDP1, we recreated the PfBDP1 knockdown in the PfAP2-I-GFP background (PfAP2-I-GFP::PfBDP1-HA-DD) (figures 7B and S7B). ChIP-qPCR assays with these parasites demonstrate that PfAP2-I is bound to the target genes (figures 7C-D) in the absence of PfBDP1 (minus Shld-1) supporting a model in which PfAP2-I binds the promoters of specific invasion genes before PfBDP1 recruitment to the same regions (figure 6).
Figure 7. PfAP2-I binding precedes PfBDP1 binding to the target gene promoters.
A- ChIP-qPCR demonstrates that PfAP2-I-GFP, but not PfBDP1-HA, is already bound to its target promoters at 22hpi. Data are represented as mean ± SD and n=2. The schizont data is the same as in Figure S6B (For PfBDP1 ChIP positive control see Figure S7A). B- In the presence of Shld-1, there are wildtype levels of PfBDP1, but in the absence of Shld-1, PfBDP1-DD is degraded and target gene transcription cannot be initiated (Josling et al., 2015). PfAP2-I may or may not remain associated to the TGCA DNA motif in the absence of PfBDP1. C- ChIP-qPCR shows that in PfAP2-I-GFP::PfBDP1-HA-DD parasites (see Figure S7), PfAP2-I-GFP remains bound to the target gene promoters even when PfBDP1 is knocked down (- Shld-1). Data are represented as mean ± SD and n=3. rh4 was used as negative control for DNA-binding.
Discussion
In Plasmodium parasites, gene transcription is highly periodic, with maximal transcript levels reached only once during the 48-hour IDC. Late in the IDC, the parasite induces the transcription of genes encoding proteins involved in RBC invasion (Bozdech et al., 2003; Le Roch et al., 2004). Two DNA elements are enriched in the promoters of invasion genes; ACAACT is present in the promoters of genes encoding micronemal and RON proteins and NGGTGCA is associated with the promoters of rhoptry bulb, msp and some glideosome genes (Elemento et al., 2007; Essien and Stoeckert, 2010; Harris et al., 2011; Iengar and Joshi, 2009; Russell et al., 2015; Young et al., 2008). Based on in vitro DNA-binding data (Campbell et al., 2010) we hypothesized that PfAP2-I binds the NGGTGCA DNA motif and drives expression of invasion-related genes.
By ChIP-seq we have shown that PfAP2-I is enriched at the promoters of invasion genes, binding to 45 genes from a previously generated “invasion list” (Bozdech et al., 2003) and 63 genes from a predicted “invadome” subnetwork (Hu et al., 2010) (Table S5). Furthermore, many of the genes bound by PfAP2-I are expressed at the mid- to late-schizont stage since 40% of the target genes are included in either a schizont-specific or an invasion-related microarray expression cluster (Le Roch et al., 2003) (Table S5). We predict that many of the PfAP2-I target genes encoding proteins of unknown function (30% of total) may also have unrecognized functions during invasion and merozoite development.
Notably, genes encoding known invasion protein complexes are co-regulated by PfAP2-I. For example, PfRAP2 interacts with PfRAP3, which is escorted to the rhoptries via interaction with PfRAP1 (Baldi et al., 2000), and is itself escorted via association with the rhoptry-associated membrane antigen (PfRAMA) protein (Richard et al., 2009). PfAP2-I binds the promoters of all four of these genes. Similarly, PfAP2-I binds upstream of pfmsp1 and pfmps7 and PfMSP1 is known to interact with PfMSP6 and PfMSP7 (Pachebat et al., 2001; Trucco et al., 2001). Genes encoding glideosome proteins PfGAP40/45/50 are also PfAP2-I targets. The absence of the GTGCA motif and lack of PfAP2-I binding to micronemal and ron gene promoters indicates that genes encoding for proteins involved in invasion stages post-RBC recognition, such as PfAMA1, RONs, EBLs or Rhs, are likely transcribed by an unidentified TF that may bind the “micronemal” ACAACT motif present upstream of these genes (Young et al., 2008). PfAP2-I also binds upstream of genes encoding the cAMP-dependent protein kinase regulatory and catalytic subunits (PKA-r and -c), which regulate microneme secretion prior to invasion (Dawn et al., 2014). Interestingly cytoadherence-linked asexual genes (CLAG) proteins, are also targets of PfAP2-I. Although the CLAGs are members of the RhopH protein family, they are not involved in invasion but their expression matches that of invasion-related genes (Counihan et al., 2017; Ito et al., 2017; Sherling et al., 2017).
Previous in vitro DNA binding experiments have predicted that PfAP2-I regulates the expression of nucleosome-related genes (Campbell et al., 2010). Our ChIP-seq data confirmed that PfAP2-I binds the promoter regions of most histone genes as well as seven apiap2 genes, including pfap2-I and five apiap2 genes that are transcribed later than pfap2-I. Among the apiap2 target genes, only PfSIP2 (Flueck et al., 2010) and PfAP2-G (Kafsack et al., 2014; Sinha et al., 2014) have been functionally characterized. A third ApiAP2 protein, PF3D7_1107800 may be part of an actin protein complex participating in nuclear repositioning of var genes (Zhang et al., 2011). The identification of other apiap2 genes as PfAP2-I targets provides evidence for ApiAP2 proteins functioning in a transcriptional regulatory network. PfAP2-I also associates with the promoters of known cell division-related genes. These include dynein -light and -heavy chain (PfDLC and PfDHC) (reviewed in (Striepen et al., 2007), cartwheel protein PfSas-6 (Suvorova et al., 2015), and MORN-repeat containing protein 1 (PfMORN1) (Heaslip et al., 2010; Lorestani et al., 2010). Genes encoding proteins involved in RBC remodeling are also targets of PfAP2-I, namely early-transcribed membrane proteins (ETRAMPs) 5 and 10.2 (Spielmann et al., 2003), and membrane-associated histidine rich protein 1 (PfMAHRP1) (Spycher et al., 2008).
The GTGCA DNA motif is highly enriched in our ChIP-seq data. PfAP2-I binding to GTGCA is important for transcription since mutation of this DNA motif upstream of msp5 affects PfAP2-I binding and gene expression (figure 3B-E), strongly indicating that PfAP2-I is the TF responsible for transcribing msp5. While binding of PfAP2-I D3 to the GTGCA DNA-sequence is essential, parasites carrying mutations that abrogate DNA binding of D1 or D2 are viable, indicating that D3 is sufficient to mediate DNA binding during the RBC stage (figure 4). We hypothesize that D3 may be a stronger DNA binder or that protein domains in proximity to the other AP2 domains influence DNA binding. Indeed PBM binding of the PfAP2-I AT-hook domain fused to D2 (AT-D2) did not recapitulate binding of D2 alone but rather that of the AT-hook (AT) (Figures S5E-F and Table S6). Post-translational modification of the AP2 domains might also alter DNA binding; D2 is acetylated during the late stages of the IDC when PfAP2-I binds the target genes and an acetylation mimic of D2 cannot bind DNA by PBM (Cobbold et al., 2016). Another possibility is that D1 and D2 are important at other stages of the life cycle; PfAP2-I is expressed in ookinetes (Lopez-Barragan et al., 2011) and its P. yoelii orthologue (PY17X_1209100) is expressed in sporozoites (Lindner et al., 2013).
Our IP experiments found that PfAP2-I co-purifies with proteins previously identified in a nuclear proteome (Oehring et al., 2012). ChIP-qPCR suggests that PfAP2-I binds to its target genes in a complex with PfCHD1 and PfBDP1. PfBDP1 has previously been shown to regulate transcription of invasion-related genes and to be required for host cell invasion. However, although PfBDP1 binds H3K9ac in vitro, this alone cannot explain how PfBDP1 is recruited to target genes as H3K9ac is much more widely distributed in the genome than is PfBDP1 (Josling et al., 2015). Our data indicate that PfAP2-I DNA-binding precedes recruitment of PfBDP1, and we speculate that PfBDP1 transcriptional activity may require interaction with TFs (such as PfAP2-I) that can directly recognize specific DNA motifs across the genome. In accordance with this model, we suggest that PfAP2-I first binds to NGGTGCA DNA motifs and recruits PfBDP1 (figure 6) and that interaction with an unidentified TF also enables PfBDP1 recruitment to the CWTAACTW DNA motif. Our results show that when PfBDP1 protein levels are knocked down, PfAP2-I remains associated to its target gene promoters, and as shown in (Josling et al., 2015) their transcript levels are reduced. We propose that PfAP2-I DNA-binding is not sufficient to induce gene transcription and requires interaction with PfBDP1. This is similar to what is seen in human cells where some TFs can only transcribe target genes when interacting with bromodomain proteins (reviewed in (Shi and Vakoc, 2014)). Such interactions occur through the association of the bromodomain with acetylated lysines in the TFs or via association of the phosphorylated bromodomain to the TFs. Intriguingly, PfAP2-I is acetylated at the trophozoite stage (Cobbold et al., 2016) and PfBDP1 is phosphorylated (reviewed in (Doerig et al., 2015)). In addition to a bromodomain, PfBDP1 also has three ankyrin repeats (Aravind et al., 2003), which may be involved in protein-protein interactions. Thus, further experimentation is required to determine how PfAP2-I binding to GTGCA DNA motifs and interaction with PfBDP1mediates transcription of invasion genes (figure 6). Overall, our data suggest that PfAP2-I plays a key role in the transcription of a subset of invasion genes by recruiting the bromodomain protein PfBDP1, and thus that PfAP2-I may be a potential therapeutic target.
STAR methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Manuel Llinás (manuel@psu.edu).
Experimental model and subject details
Parasite culture and transfection
P. falciparum parasites were cultured at 37C in the presence of 5% oxygen and 7% carbon dioxide, in RPMI 1640 media supplemented with hypoxanthine and 0.5% Albumax II (Invitrogen) (culture media recipe can be found at llinaslab.psu.edu/protocols/) in 2% or 5% haemotocrit. Parasites were fed fresh blood every other day and parasitemia was kept at around 2–5% by counting the number of parasites in blood smears (Trager and Jensen, 1976).
Parasite synchronization to the ring stage was done by incubating pelleted cultures 1:1 with 0.3M L-Alanine, 10mM Hepes pH 7.5 solution for 10 minutes at 37C, after which cultures were centrifuged, resuspended in fresh media and placed back in culture (Braun-Breton et al., 1988). All experiments, unless stated otherwise, were performed at 40h post-invasion.
When necessary to induce a knockdown, DD-FKBP parasites were synchronized and Shld-1 was removed for 26h.
Parasite strains
| Dd2 | wt strain | grown in standard culture media |
| PfAP2-I-GFP | Dd2 strain with endogenous PfAP2-I C-terminally tagged with GFP | grown in standard media supplemented with BSD |
| PfSIP2N-HA | 3D7 strain expressing episomal copy of PfSIP2 N-terminus fused to 3xHA | grown in standard media supplemented with BSD (parasite line previously generated) |
| PfAP2-I-GFP::msp5MUT | PfAP2-I-GFP Dd2 strain with mutated PfAP2-I DNA motif in the promoter of endogenous msp5 | grown in standard media |
| PfAP2-I-GFP-D1mut | PfAP2-I-GFP Dd2 strain with mutated AP2 domain 1 (D1) | grown in standard media |
| PfAP2-I-GFP-D2mut | PfAP2-I-GFP Dd2 strain with mutated AP2 domain 2 (D2) | grown in standard media |
| PfAP2-INLS-GFP | Dd2 strain expressing episomal copy of NLSPfAP2-I-GFP | grown in standard media supplemented with WR |
| NLSPfSIP2-GFP | Dd2 strain expressing episomal copy of NLSPfSIP2-GFP | grown in standard media supplemented with WR |
| NLSCalmodulin-YFP | 3D7 strain expressing episomal copy of NLSCalmodulin-YFP | grown in standard media supplemented with WR (parasite line previously generated) |
| HDGFP | 3D7 strain expressing episomal copy of NLSCalmodulin-YFP | grown in standard media supplemented with WR (parasite line previously generated) |
| PfAP2-I-GFP::PfBDP1-HA-DD | PfAP2-I-GFP Dd2 strain with endogenous PfBDP1 C-terminally tagged with 3xHA and FKBP (DD) | grown in standard media supplemented with WR and BSD |
All parasite strains are available upon request.
Bacterial strains
| pB-3’pfap2-i-GFP | DH5α bacterial strain carrying pB-3’pfap2-i-GFP |
| pJDD-5’bdp1 | DH5α bacterial strain carrying pJDD-5’bdp1 |
| pDC2-cam-PfAP2-INLS-GFP | DH5α bacterial strain carrying pDC2-cam-PfAP2-INLS-GFP |
| pDC2-CAM-PfSIP2NLS-GFP | DH5α bacterial strain carrying pDC2-CAM-PfSIP2NLS-GFP |
| pL7-msp5MUT-gRNAmsp5 | DH5α bacterial strain carrying pL7-msp5MUT-gRNAmsp5 |
| pL7-D1MUT-gRNAD1 | DH5α bacterial strain carrying pL7-D1MUT-gRNAD1 |
| pL7-D2MUT-gRNAD2 | DH5α bacterial strain carrying pL7-D2MUT-gRNAD2 |
| pL7-D3MUT-gRNAD3 | DH5α bacterial strain carrying pL7-D3MUT-gRNAD3 |
| pGEX-D1-GST | BL21 bacterial strain carrying pGEX-D1-GST (previously generated) |
| pGEX-D2-GST | BL21 bacterial strain carrying pGEX-D2-GST (previously generated) |
| pGEX-D1mut-GST | DH5α bacterial strain carrying pGEX-D1mut-GST |
| pGEX-D1mut-GST(BL21) | BL21 bacterial strain carrying pGEX-D1mut-GST |
| pGEX-D2mut-GST | DH5α bacterial strain carrying pGEX-D2mut-GST |
| pGEX-D2mut-GST(BL21) | BL21 bacterial strain carrying pGEX-D2mut-GST |
| pGEX-D3mut-GST | DH5α bacterial strain carrying pGEX-D3mut-GST |
| pGEX-D3mut-GST(BL21) | BL21 bacterial strain carrying pGEX-D3mut-GST |
All DH5α strains were grown in LB media supplemented with Ampicillin and all BL21-CodonPlus(DE3)-RIL cells (BL21) strains were grown in LB media supplemented with chloramphenicol. All bacterial strains are available as glycerol stocks upon request.
Method details
P. falciparum Transfections
Plasmid transfections into P. falciparum parasites were performed by electroporation using a Gene-Pulser II (Bio-Rad) as previously described (Fidock and Wellems, 1997). Briefly, ring-stage parasites at 5% haemotocrit were washed 3 times with warm cytomix (120 mM KCl, 0.2 mM CaCl2, 2 mM EGTA, 10 mM MgCl2, 25 mM HEPES, 5 mM K2HPO4, 5 mM KH2PO4; pH 7.6) and resuspended to 50% haemotocrit with ice-cold cytomix. Parasites were then electroporated with 100µg of plasmid DNA resuspended in cytomix. All CRISPR/Cas9 parasite lines were generated by co-transfecting parasites with 30µg of pL7 plasmid and 30µg of the pUF1-Cas9 plasmid (Ghorbal et al., 2014). Transfected parasites were resuspended in warm media and cultured overnight in complete media plus 2% hematocrit. The next day, transfectants were selected on either 2µg/ml blasticidin-S-HCl (BSD) and/or 2.5nM WR99210 (WR) and/or 1.5µM 5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)naphthalen-2-ylamine (DSM1). PfAP2-I-GFP::PfBDP1-HA-DD parasites were generated by growing parasites in the presence of 0.5µM Shld-1 (Takara). When PfAP2-I-GFP Dd2 parasites were used for transfection, the media was supplemented with BSD. When necessary, parasites were cycled ON/OFF drugs for at least 2 cycles and cloned by serial dilution in order to ensure that the population was clonal for plasmid integration. Transfected lines were verified by PCR using genomic DNA collected with a DNeasy Blood & Tissue kit (Qiagen, RRID: SCR_008539) and using the appropriate primers and by Western-blot. Parasite lines generated using CRISPR/Cas9 had the locus of interest sequenced by Sanger sequencing.
Antibodies
The primary antibodies used were: mouse anti-GFP 11814460001 (Roche, RRID: AB_390913) (Western-blot, 1:1000 dilution), in-house developed rabbit anti-GFP (Cristea et al., 2005) (IP), mouse anti-Histone H3 10799 (Abcam, RRID: AB_470239) (Western-blot, 1:2000 dilution), rabbit anti-aldolase 38905 (Abcam, RRID: AB_771788) (Western-blot, 1:1000 dilution), rat anti-HA high affinity 3F10 (Roche, RRID: AB_10094468) (Western blot: 1/200 dilution and ChIP), rabbit anti-GFP 290 (Abcam, RRID: AB_303395) (ChIP), rabbit anti-IgG 46540 (Abcam, RRID: AB_2614925) (ChIP and IP), rabbit anti-MSP4 MRA-319 (MR4) (Western-blot, 1:1000 dilution), and rabbit anti-MSP5 MRA-320 (MR4) (Western-blot, 1:1000 dilution),. Secondary antibodies used were: goat anti-mouse IgG (Fc) peroxidase conjugated (ThermoFisher Scientific), goat anti-rabbit IgG HRP-conjugate (Millipore, RRID: AB_390191) and goat anti-rat IgG HRP-conjugate (Millipore, RRID: AB_11214444) at 1:3000 dilutions. Hoechst 33342 (Life Technologies) was used in all live immunofluorescence assays to visualize the nucleus. For details on the concentration of antibodies used for ChIP or IP refer to those protocols below.
Plasmid DNA cloning
All primers used for cloning are listed in Table S1.
Restriction enzymes were purchased from New England Biolabs (NEB, RRID:SCR_013517).
All PCRs were performed using Advantage Genomic LA Polymerase Mix (Clontech) according to the manufacturer’s instructions.
In order to tag pf3d7_1007700 (pfap2-i) with GFP, 1040bp from the 3’end of the gene were amplified from 3D7 genomic DNA with primers AP2-I-1 and AP2-I-2 (Table S1) and cloned into the pBcam-GFP plasmid between BglII and NotI, generating pB-3’pfap2-i-GFP. The plasmid pBcam-GFP was generated by amplifying the GFP sequence with primers GFP-1 and GFP-2 and cloning into the pBcam-3xHA plasmid (Flueck et al., 2009) between NotI and SalI.
To endogenously tag pfbdp1 with 3xHA-DD-FKBP in PfAP2-I-GFP parasites, 1383bp of the 3’end of the gene was amplified from 3D7 genomic DNA with primers BDP1-1 and BDP1-2 and cloned into the pJDD41 plasmid (Dvorin et al., 2010) between NotI and XhoI, generating pJDD41-3’bdp1.
In order to express the putative NLS of PfAP2-I or PfSIP2 fused to GFP, primers NLS2-1 and NLS2-2 or SIP2-1 and SIP2-2 were used to amplify the NLS sequences and the PCR products were cloned into the pDC2-cam-CRT-GFP plasmid (Fidock et al., 2000) between AvrII and SpeI, generating pDC2-cam-PfAP2-INLS-GFP and pDC2-CAM-PfSIP2NLS-GFP, respectively.
The ATGCA DNA motif upstream of msp5 was mutated in PfAP2-I-GFP parasites using the plasmid pL6-msp5MUT-gRNAmsp5, which was generated by cloning the guide RNA created by annealing the primers gRNA MSP5-1 and gRNA MSP5-2 and cloning the annealed product into the pL6-CS plasmid (Ghorbal et al., 2014) into the BtgZI site using In-Fusion HD Cloning Plus (Clontech) according to the manufacturer’s instructions.
The msp5MUT sequence was generated by amplifying two mutated arms with primers MSP5-1 and MSP5-2 and primers MSP5-3 and MSP5-4, then ligating with In-Fusion (primers MSP5-2 and MSP5-3 have 15bp homology with each other) and re-amplifying the ligated product with primers MSP5-1 and MSP5-4 (Table S1). The final mutated product was cloned with In-Fusion into the pL6-gRNAmsp5 plasmid between NotI and SpeI (primers MSP5-1 and MSP5-4 have 15bp homology to the pL6-CS plasmid), generating pL7-msp5MUT-gRNAmsp5. The plasmids used to mutate the first, second and third AP2 domains of PfAP2-I in PfAP2-I-GFP parasites were created by cloning the guide RNA created by annealing the primers gRNA D1-1 and gRNA D1-2 or gRNA D2-1 and gRNA D2-2 or gRNA D3-1 and gRNA D3-2, respectively, and cloning the annealed products In-Fusion into the pL6-CS plasmid in the BtgZI site, according to the manufacturer’s instructions. The D1MUT and D2MUT sequences were generated by amplifying mutated arms with primers D1-1 and D1-2 and primers D1-3 and D1-4 or with primers D2-1 and D2-2 and primers D2-3 and D2-4 (Table S1), then ligating with In-Fusion and re-amplifying the ligated product with primers D1-1 and D1-4 or D2-1 and D2-4. The final mutated products were cloned into the pL6-gRNAD1 or pL6-gRNAD2 plasmids between NotI and SpeI, generating pL7-D1MUT-gRNAD1 and pL7-D2MUT-gRNAD2, respectively. The D3MUT sequence (2727-3100bp pfap2- I ORF) was gene synthesized by Genewiz (RRID:SCR_003177) (Table S1) and cloned into pL7-gRNAD3 between Not I and SpeI, generating pL7-D3MUT-gRNAD3.
DNA sequences encoding for the AT-hook alone (AT) or the AT-hook fused to the second AP2 domain (AT-D2) were PCR amplified from 3D7 gDNA using primers AT-1 and AT-2 or AT-1 and D2-5, respectively, and cloned into the BamHI and XhoI sites in the pGEX-4T1 plasmid (GE Life Sciences, RRID:SCR_000004) to create the N-terminal glutathione S-transferase (GST) fusions pGEX-AT-GST and pGEX-AT-D2-GST. DNA sequences encoding mutant versions of the first (D1), second (D2) or third (D3) AP2 domains were PCR amplified from pL7-D1MUT-gRNAD1, pL7-D2MUT-gRNAD2 or pL7-D3MUT-gRNAD3 using primers D1-4 and D1-5 or D2-6 and D2-7 or D3-1 and D3-2, respectively, and cloned into the BamHI and XhoI sites in the pGEX-4T1 plasmid to create the N-terminal glutathione S-transferase (GST) fusions pGEX-D1mut-GST, pGEX-D2mut-GST and pGEX-D3mut-GST.
All plasmids were verified by Sanger-sequencing and diagnostic restriction digestion.
Expression and purification of recombinant proteins
GST-tagged recombinant proteins were expressed in BL21-CodonPlus(DE3)-RIL cells (Agilent Technologies) by inducing with 0.2mM IPTG at room temperature overnight, and were affinity purified using glutathione resin (Clontech) (protocol can be found at llinaslab.psu.edu/protocols/). The efficiency of the purification was estimated by Coomassie blue staining. GST-D1 proteins could not be purified and the bacterial lysate was used instead for protein binding microarrays.
Protein Binding Microarrays
Bacterial lysate or purified recombinant proteins were assayed for DNA binding as previously described in (Campbell et al., 2010) using protein binding microarrays (PBMs). Briefly, custom designed DNA oligonucleotide arrays were double-stranded using a universal primer, incubated with GST-AP2 fusion proteins, visualized with Alexa-488 conjugated anti-GST antibody (Millipore, RRID:AB_2534137), and scanned using an Axon 4200A scanner. Proteins were used at the maximum concentration obtained from purification and represent one-fifth of the total reaction volume used on the PBM. After data normalization and calculation of enrichment scores the “Seed-and-Wobble” algorithm was applied to combine the data from two separate experiments and create position weight matrices (PWMs).
As a control, PBMs were performed with the lysate of GST-D1 expressing cells or newly purified GST-D2 protein previously used in (Campbell et al., 2010). Each GST-tagged protein was run on two PBM array versions, AMADID 016060 (v9) and AMADID 015681 (v11), with the exception of GST-AT which was only tested on v9. DNA binding motifs from the two arrays were combined to generate the final binding motif reported.
Bioinformatic searches
Simple Modular Architecture Research Tool (SMART) (Letunic et al., 2012; Schultz et al., 1998) was used to predict the AT-hook motif in the PfAP2-I sequence. NLS were predicted using NLStradamus (Nguyen Ba et al., 2009). Multalin (Corpet, 1988) was used for the protein sequence alignments shown throughout.
Nuclear fractionation assays and Western blot
Nuclear fractionation assays were performed as previously described (Flueck et al., 2010). Briefly, saponin-lysed infected red blood cells were lysed in ice-cold cell lysis buffer (20mM Hepes pH 7.9, 10mM KCl, 1mM EDTA, 1mM EGTA, 1mM DTT, protease inhibitors) for 5 min. After a 5 min centrifugation at 5200 rpm (2500xg), the cytosolic extract was collected and the cell pellet was re-suspended in ice-cold nuclear extraction buffer (20mM Hepes pH 7.9, 1M KCl, 1mM EDTA, 1mM EGTA, 1mM DTT, protease inhibitors). After a 30 min incubation at 4C, the mixture was centrifuged at 11800 rpm (13000xg) for 30 min and the nuclear extract collected (protocol can be found at llinaslab.psu.edu/protocols/). The cytosolic and nuclear extracts were diluted in SDS-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer containing 100mM DTT and boiled for 10 min before running in a SDS-PAGE gel. SDS-PAGE was performed using standard methods. Separated proteins were transferred to a nitrocellulose membrane and western blots were performed using primary antibodies diluted in 5% non-fat milk powder in 1X PBS-0.05% Tween. As a secondary antibody, a peroxidase-conjugated goat anti-mouse/rabbit/rat antibody was used. Bound antibodies were visualized using an established ECL Western blotting substrate (Pierce).
Immunoaffinity purification assays
High salt nuclear extracts from PfAP2-I-GFP parasites were prepared as described above for the nuclear fractionation assays. The cytosolic and nuclear extracts were then diluted 4 times in dilution buffer (20mM Hepes pH7.9, 1mM EDTA, 1mM EGTA, 40% glycerol, protease inhibitors).
5mg M-270 Epoxy Dynabeads (ThermoFisher Scientific) were conjugated with 12.5µg of a custom polyclonal anti-GFP antibodies (Cristea et al., 2005) or 25µg IgG. Beads were twice washed in 0.1M sodium phosphate buffer (pH 7.4) and conjugated by incubating the beads with the antibody (20µl total volume per milligram of beads), 1M ammonium sulfate and 0.1M sodium phosphate overnight at 30C, while rotating the tubes. After extensive washing, the conjugated beads were stored at 4C in 1xPBS supplemented with 0.02% NaN3 for up to one week (Joshi et al., 2013).
The diluted nuclear and cytosolic extracts were incubated with the conjugated beads as previously described in (Joshi et al., 2013) for 1.5h, at 4C. The beads were then extensively washed, and the protein complexes were eluted and digested in-solution with trypsin. The eluates were mixed with 8M Urea buffer in an ultra-filtration device and centrifuged 30min. The filters were then first washed once with 1M iodoacetamide and then three times with 0.01M ammonium bicarbonate. The washed filter units were transferred into a tube pre-washed with 200µl 50% acetonitrile, and the peptides were digested with 100 µl of 5ng/µl trypsin at 37C, overnight. The next day, the filter units were centrifuged and eluted twice with 50µl HPLC water. The elutions were transferred into an autosampler vial pre-loaded with 22µl of 10% trifluoroacetic acid and the samples were concentrated by vacuum centrifugation until the total volume was 10–30µl (Greco et al., 2012). The samples were then analyzed by mass spectrometry.
The immunoaffinity purification (IP) eluted material was digested using the filter aided sample preparation (FASP) protocol (Wisniewski et al., 2009). Enzymatic digestion was performed in 100 ml of trypsin solution (5 ng/ml in 100mM ammonium bicarbonate) in Vivacon 500 centrifugal filters (10 kDa MWCO; Sartorius Stedim Biotech, Goettingen, Germany). Digested peptides were concentrated by vacuum centrifugation, desalted using StageTips constructed using Empore C18 extraction discs (3M Analytical Biotechnologies). Alternatively, the eluted material obtained from the immunopurification done with PfAP2-I-GFP parasites and GFP-conjugated beads underwent gel-assisted proteolysis, as described in (Han et al., 2008) (Table S4). Desalted peptides were analyzed by nanoliquid chromatography–tandem mass spectrometry using a Dionex Ultimate 3000 nRSLC coupled to an LTQ-Orbitrap Velos mass spectrometer (ThermoFisher Scientific, San Jose, CA), as described (Joshi et al., 2013). MS/MS spectra were extracted by Proteome Discoverer and analyzed using SEQUEST (Eng et al., 1994) by searching P. falciparum and contaminant protein databases.
As controls, extracts from parasites expressing GFP fused to nuclear localization signals (NLS) - NLSPfSIP2-GFP (figure S6A) or NLSCalmodulin-YFP (Russo et al., 2009) – or expressing GFP in the cytosol – HDGFP (Kadekoppala et al., 2000) were also prepared (Figure S1C and Table S4). Each immunoaffinity purification was performed two independent times and only proteins that were detected in both replicates with 3 or more peptides were considered for further analysis.
DNA Microarrays
RNA from tightly synchronized PfAP2-I-GFP and PfAP2-I-GFP::msp5MUT parasites was harvested every 6h starting at 42h post-invasion for 24h (Figure S3G) and purified using a TRIzol (ThermoFisher Scientific) extraction method. The RNA was then reverse transcribed into cDNA using Superscript II RT (ThermoFisher) according to the manufacturer’s instructions, and the cDNA was concentrated using DNA Clean and Concentrator-5 (Zymo Research) according to the manufacturer’s instructions. The cDNA was labeled with CyDye (Cy3/Cy5) using Amersham CyDye Reactive Dyes (GE Healthcare) according to the manufacturer’s instructions, and purified using DNA Clean and Concentrator- 5 kit. DNA microarrays were performed as described in (Painter et al., 2013). RNA extraction, reverse transcription, cDNA labeling, array hybridization and washing protocols can be found at llinaslab.psu.edu/protocols/.
ChIP-seq
The ChIP-seq assay was performed as described in (Lopez-Rubio, 2012) with some modifications. Synchronized 40h schizont stage parasite cultures were either formaldehyde-crosslinked and then lysed with saponin until complete red blood cell lysis (ChIP replicates 1 and 2), or saponin-lysed and then formaldehyde-crosslinked (ChIP replicate 3). The isolated chromatin was sheared in SDS lysis buffer to obtain a fragment size of 100–150bp using an M220 focused-ultrasonicator (Covaris Inc.) and the following settings: peak power 75W, 2% duty factor, 200 cycles per burst and total treatment time of 900s. After pre-clearing the chromatin with Magna ChIP protein A+G magnetic beads (Millipore) (ChIP replicates 1 and 2) or protein A salmon sperm agarose beads (Millipore) (ChIP replicate 3), an input sample was collected and the rest of the chromatin was incubated overnight at 4C with 1µg of polyclonal anti-GFP or, as control, the same amount of IgG. The immunoprecipitated chromatin was collected with A+G magnetic beads (ChIP replicates 1 and 2) or agarose beads (ChIP replicate 3), extensively washed and eluted with elution buffer. The input and ChIP samples were reverse cross-linked overnight at 45C in the presence of 0.4M NaCl and purified by phenol:chloroform or using the Qiagen MinElute PCR purification kit.
Barcoded libraries for Illumina TruSeq single-end sequencing were constructed using NEBNext DNA library Prep reagents (New England Biolabs) by following the standard Illumina library preparation protocol. The DNA was first end-repaired for 30 min at 20C, purified using Agencourt AMPure XP beads (Beckman Coulter) (ChIP replicates 1 and 2) or via phenol:chloroform extraction (ChIP replicate 3) and then dA-tailed for 30 min at 37C, and purified using Ampure XP beads (ChIP replicates 1 and 2) or via phenol:chloroform extraction (ChIP replicate 3). NEXTflex Illumina DNA barcodes (diluted 1/10) were then ligated to the DNA fragments using T4 DNA ligase at 20C for 15 mins. The resulting DNA was then size selected for 250 bp inserts using Ampure XP beads (ChIP replicates 1, 2 and 3). Afterward the libraries were PCR-amplified using Kapa HiFi (Kapa biosystems) and purified using AMPure XP beads. The quality and percentage of adaptor-ligated material of the final sequencing libraries was determined by running them on an Agilent 2100 Bioanalyzer (Agilent Technologies). The concentration of each library was determined using a Qubit dsDNA HS Assay kit (Invitrogen). The final libraries were multiplexed with three to fourteen barcoded samples and contained up to 20% PhiX control DNA per lane on an Illumina (RRID:SCR_010233) HiSeq 2500 system to generate 150 base pair single-end reads.
ChIP-qPCR
The primers used for ChIP-qPCR are listed in Table S1.
The ChIP-qPCR assays were performed as described for ChIP-Seq with few exceptions. The isolated chromatin was sheared in SDS lysis buffer to obtain a fragment size of 500bp using an M220 focused-ultrasonicator and the following settings: 10% duty factor, 200 cycles per burst and total treatment time of 60s. After pre-clearing, an input sample was collected and the rest of the chromatin was incubated overnight at 4C with 1µg of polyclonal anti-GFP/anti-HA antibody or, as control, the same amount of IgG. The immunoprecipitated chromatin was collected with Magna ChIP protein A+G magnetic beads (Millipore), extensively washed and eluted with elution buffer. The input and ChIP samples were reverse cross-linked overnight at 45C in the presence of 0.4M NaCl, and purified by phenol:chloroform extraction or using the Qiagen MinElute PCR purification kit. For quantitative PCR (qPCR), the concentration of each eluted sample was determined by Qubit dsDNA Broad-Range Assay Kit (Invitrogen), diluted 10 times and used for quantitative PCR in triplicate wells using PowerSYBR Green Master Mix (ThermoFisher Scientific) as described (Crowley et al., 2011). Each ChIP-qPCR experiment was performed with at least 3 biological replicates.
For ChIP-qPCR from the PfAP2-I-GFP::PfBDP1-HA-DDline, parasites were grown +/− Shld-1 for the times indicated and, after pre-clearing, an input sample was collected. The remainder of the chromatin was incubated overnight at 4C with 1µg of polyclonal anti-GFP or anti-HA antibody. As a control, the same amount of IgG antibody was used.
RT-qPCR
cDNA was synthesized from approximately 2µg RNA using SuperScript II Reverse Transcriptase (ThermoFisher Scientific) with random nonamers and oligo(dT) primers according to the manufacturer’s instructions. Controls containing all reaction components except SuperScript II were also set up to serve as no enzyme controls. qPCR was performed as described above. The list of primers used is in Table S1.
Quantification and statistical analysis
Western-blot protein quantification
Protein quantification was performed using the ImageLab software of ChemiDoc MP Imaging System (Bio-Rad).
Nuclear extraction efficiency was estimated using anti-histone H3 and anti-aldolase as nuclear and cytosolic markers, respectively.
ChIP-qPCR and RT-PCR data analysis
ChIP-qPCR and RT-PCR data were analyzed using the ΔΔCt method.
3D7 wt parasites or in the case of the parasite line PfAP2-I-GFP::PfBDP1-HA-DD, parasites treated without the ligand Shld-1, were used as controls.
pf3d7_0717700 (serine tRNA ligase) was used as a reference gene for RT-PCR.
In all figure legends, n denotes biological replicates (independently collected samples). Data are represented as mean ± SD.
DNA Microarrays
All sample data was compared to an arbitrary reference pool made-up of a mixed parasite population of rings, trophozoites and schizonts. The results shown represent the log2 ratio against the cDNA reference pool and are the average of 2 biological replicates.
Rnits (Sangurdekar, 2014) was used to determine a p-value for all transcripts detected by DNA microarray (minus antigenic variant genes). Rnits compares multiple time-series expression data sets by summarizing probes into gene-level information. For all genes, it fits a series of B-splines with varying curvature and degrees of freedom. Under the null hypothesis H_0, a single model is fit for all data sets. P-values from the hypothesis test are then plotted and the least complex spline parameters that result in uniformly distributed null p-values are automatically chosen. Each gene is attributed a p-value from 0 to 1 until all p-values are uniformly distributed. While genes with p-values closer to 1 have fewer changes in transcription levels between expression data sets, genes with p-values closer to 0 have higher transcription levels differences between time-series expression data sets.
Protein binding microarrays
An enrichment score greater than 0.45 indicates high affinity binding in PBM experiments. Although the combined v9 and v11 results for the GST-D2wt control scored below 0.45, the individual v9 PBM had an E-score of 0.46 and the same binding motif was reported previously (Campbell et al. 2010). The score for each 8-mer nucleotide sequence reflects the affinity of a DNA binding domain for that sequence, with higher scores representing tighter interactions.
ChIP-seq data analysis
ChIP-seq reads from 3 biological replicates were demultiplexed and then processed using FastQC (Andrew, 2010) (version 0.11.2, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Trimmomatic (Bolger et al., 2014). Trimming was performed by removing low quality bases and adapters using a minimum sliding window length and quality of 4 and 30, respectively, and removing reads below 36bp. Trimmed reads were mapped to the P. falciparum Dd2 draft genome (ftp://ftp.sanger.ac.uk/pub/pathogens/Plasmodium/falciparum/PF3K/PilotReferenceGenomes/DraftAnnotation/PfDd2/) using BWA MEM (Li, 2013), version 0.7.13-r1126, and the default parameters. SAMtools version 1.3 (Li et al., 2009) was used to remove multi-mapped reads, non-primary alignments and PCR duplicates. Peaks were called using MACS2 version 2.1.1.20160309 (Zhang et al., 2008) callpeak function and a q-value cutoff of 0.05 (further details in supplemental experimental procedures). Only peaks that were detected in at least 2 of the 3 biological replicates were considered for further analysis; 177 trimmed peaks were further analyzed.
Anti-GFP and anti-IgG ChIP-seq libraries were compared and normalized to their respective input libraries using MACS2 for each individual replicate. Peaks within anti-IgG samples were used to filter anti-GFP libraries to remove false positives using the BEDTools suite (Quinlan and Hall, 2010) and pybedtools library (Dale et al., 2011). Peaks for each replicate were analyzed individually for technical consistency and a final list of peaks was computed by determining the overlap between biological replicates using the BEDTools multiinter command. These “trimmed” peaks represent intervals where peaks from biological replicate experiments overlapped. Only “trimmed” peaks present in at least 2/3 biological replicates were considered for further analysis.
Motif enrichment analysis of the trimmed peaks was done using DREME (Bailey, 2011) to identify enriched motifs between 6 and 10bp as compared to random P. falciparum genomic intervals of similar length. FIMO (Grant et al., 2011) was used to search 250bp upstream and downstream of peak summits for the presence of enriched sequence motifs, as found by DREME, using a p-value threshold of 1e-3. Finally, TOMTOM (Gupta et al., 2007) was used to compare the de novo discovered motifs to previously in silico discovered motifs (Campbell et al., 2010); a match required a minimum overlap of 4 positions.
To build a motif heatmap (figure 2A), the most highly enriched motif found by DREME was used to search the trimmed peak sequences. This search was done using FIMO and a threshold of 1e-3. The top-scoring motif within each sequence was assigned to its respective peak. The sequence of each peak summit was centered on these assigned motifs and ordered based on highest scoring motif according to FIMO. The position weight matrix of the aligned heatmap was obtained using the seqLogo package (Bembom, 2017). To search for enriched motifs within the peak sequences of PfBDP1 obtained from (Josling et al., 2015), we took an analogous approach, using similar thresholds and parameters.
To annotate peaks, peak summits corresponding to peaks found in at least 2/3 ChIP replicates were used to calculate the distance to target genes. Gene annotations were assigned based on their distance to the nearest downstream gene. Alignment tracks for visualization were generated using deepTools (Ramirez et al., 2014) and were visualized using the Integrated Genomics Viewer (Thorvaldsdottir et al., 2013). Enrichment heatmaps and profile plots were generated using the deepTools computeMatrix and plotHeatmap tools. Gene Ontology (GO) enrichment analysis was done using the GO feature at PlasmoDB.org (Aurrecoechea et al., 2009).
Immunoaffinity purification
Each immunoaffinity purification (sample and controls) was performed two independent times and only proteins that were detected in both replicates with 3 or more peptides were considered for further analysis.
The data generated was analyzed and filtered using the algorithm SAINT, which attributes a pSAINT value as a probability of interaction for each protein based on the number of spectra detected on the sample (PfAP2-I-GFP IPs) versus controls IPs (Choi et al., 2011).
The final list of interacting proteins was assembled by comparing the list of peptides obtained for both methods of proteolytic digestion, as described above, including only proteins with a pSAINT value (probability of interaction) equal to or above 0.9.
Data and software availability
ChIP-seq data
All next generation sequencing data has been deposited at the NCBI Sequence Read Archive (GSE80293).
DNA Microarray data
All DNA microarray data has been deposited at the NCBI Gene Expression Omnibus (GEO) as datasets GSE77807 and GSE77810.
Protein data
All immunoprecipitation proteomics data is available in supplementary Table S4.
Protein Binding Microarray data
All PBM array data has been deposited at the UniPROBE (Universal PBM Resource for Oligonucleotide Binding Evaluation) database under accession number UP01415 (Hume et al., 2015). See Table S6 for position weight matrices for all AP2 domains tested by PBM.
Supplementary Material
Highlights.
PfAP2-I binds a DNA motif enriched upstream of merozoite invasion genes.
PfAP2-I binding to the invasion DNA motif is necessary for target gene transcription.
Only the C-terminal AP2 domain of PfAP2-I is essential for asexual stage activity.
PfAP2-I and PfBDP1 Co-IP and regulate transcription of many of the same genes.
Acknowledgments
JMS is a recipient of Swiss National Fund GEP3-13613 and EMBO Long-Term Fellowship 633-2011, TLC was funded by an NSERC Postdoctoral Fellowship, JP was funded by an NJCCR postdoctoral fellowship and GJ is the recipient of an American Heart Association Postdoctoral Research Grant. This work was funded by NIH/NIAID R01AI076276 (ML) and R01AI125565 (ML), the Arnold and Mabel Beckman Foundation, support from the Center for Quantitative Biology (P50 GM071508) (ML), and R01GM114141 (IMC). We thank the Voss (SIP2N-HA), Cowman (CHD1-GFP) and Duffy (BDP1-HA) labs for sharing parasite strains. We thank Dr. T. Greco in the Cristea lab for the mass spectrometric analysis at Princeton University and Dr. T. Laremore for those performed at the Pennsylvania State University in the Huck Proteomics and Mass Spectrometry Core Facility. We thank Drs. T. Otto, M. Berriman and U. Boehme from the UK Wellcome Trust Sanger Institute for use of the Pf Dd2 draft annotations from the Pf3K project. We thank Dr. H. Painter for help with Rnits, discussions of the data and critical reading of the manuscript. We thank A. Minns for assistance with protein quantification. High-throughput sequencing analyses were preformed at the Huck Genomics Core Facility, University Park, PA. The anti-MSP4 and anti-MSP5 antibodies were obtained through the MR4 as part of the BEI resources repository.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.M.S. and M.L conceived the experiments and wrote the manuscript. T.C. generated the PfAP2-I-GFP parasite line and initiated the project. I.M.C. contributed protocols and reagents to the IP proteomic studies and P.J. performed and analyzed some of the IP experiments. A.S. performed PBMs, L.A. ran DNA microarrays and PBMs, G.J. performed RT-qPCR and ChIP assays and P.R. handled all bioinformatic data analysis. J.M.S. performed all other experiments. All authors have read the manuscript.
References
- Andrew S. FASTQC: a quality control tool for high throughput sequence data. 2010 http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: genomic gleanings. Cell. 2003;115:771–785. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- Armstrong CM, Goldberg DE. An FKBP destabilization domain modulates protein levels in Plasmodium falciparum. Nature Methods. 2007;4:1007–1009. doi: 10.1038/nmeth1132. [DOI] [PubMed] [Google Scholar]
- Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:539–543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27:1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji S, Babu MM, Iyer LM, Aravind L. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Research. 2005;33:3994–4006. doi: 10.1093/nar/gki709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi DL, Andrews KT, Waller RF, Roos DS, Howard RF, Crabb BS, Cowman AF. RAP1 controls rhoptry targeting of RAP2 in the malaria parasite Plasmodium falciparum. EMBO Journal. 2000;19:2435–2443. doi: 10.1093/emboj/19.11.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembom O. R package version 1.42.0. Bioconductor. 2017. seqLogo: Sequence logos for DNA sequence alignments. [Google Scholar]
- Berger MF, Bulyk ML. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nature Protocols. 2009;4:393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff E, Vaquero C. In silico and biological survey of transcription-associated proteins implicated in the transcriptional machinery during the erythrocytic development of Plasmodium falciparum. BMC Genomics. 2010;11:34. doi: 10.1186/1471-2164-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biology. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun-Breton C, Rosenberry TL, da Silva LP. Induction of the proteolytic activity of a membrane protein in Plasmodium falciparum by phosphatidyl inositol-specific phospholipase C. Nature. 1988;332:457–459. doi: 10.1038/332457a0. [DOI] [PubMed] [Google Scholar]
- Bullen HE, Tonkin CJ, O'Donnell RA, Tham WH, Papenfuss AT, Gould S, Cowman AF, Crabb BS, Gilson PR. A novel family of Apicomplexan glideosome-associated proteins with an inner membrane-anchoring role. Journal of Biological Chemistry. 2009;284:25353–25363. doi: 10.1074/jbc.M109.036772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell TL, De Silva EK, Olszewski KL, Elemento O, Llinas M. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Pathogens. 2010;6:e1001165. doi: 10.1371/journal.ppat.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras AC, Nesvizhskii AI. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nature Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold SA, Santos JM, Ochoa A, Perlman DH, Llinas M. Proteome-wide analysis reveals widespread lysine acetylation of major protein complexes in the malaria parasite. Scientific Reports. 2016;6:19722. doi: 10.1038/srep19722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combe A, Giovannini D, Carvalho TG, Spath S, Boisson B, Loussert C, Thiberge S, Lacroix C, Gueirard P, Menard R. Clonal conditional mutagenesis in malaria parasites. Cell Host Microbe. 2009;5:386–396. doi: 10.1016/j.chom.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Research. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counihan NA, Chisholm SA, Bullen HE, Srivastava A, Sanders PR, Jonsdottir TK, Weiss GE, Ghosh S, Crabb BS, Creek DJ, et al. Plasmodium falciparum parasites deploy RhopH2 into the host erythrocyte to obtain nutrients, grow and replicate. eLife. 2017;6 doi: 10.7554/eLife.23217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman AF, Berry D, Baum J. The cellular and molecular basis for malaria parasite invasion of the human red blood cell. The Journal of Cell Biology. 2012;198:961–971. doi: 10.1083/jcb.201206112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Molecular & Cellular Proteomics. 2005;4:1933–1941. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- Dale RK, Pedersen BS, Quinlan AR. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics. 2011;27:3423–3424. doi: 10.1093/bioinformatics/btr539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Hertrich N, Perrin AJ, Withers-Martinez C, Collins CR, Jones ML, Watermeyer JM, Fobes ET, Martin SR, Saibil HR, et al. Processing of Plasmodium falciparum Merozoite Surface Protein MSP1 Activates a Spectrin-Binding Function Enabling Parasite Egress from RBCs. Cell Host Microbe. 2015;18:433–444. doi: 10.1016/j.chom.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawn A, Singh S, More KR, Siddiqui FA, Pachikara N, Ramdani G, Langsley G, Chitnis CE. The Central Role of cAMP in Regulating Plasmodium falciparum Merozoite Invasion of Human Erythrocytes. PLoS Pathogens. 2014;10:e1004520. doi: 10.1371/journal.ppat.1004520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerig C, Rayner JC, Scherf A, Tobin AB. Post-translational protein modifications in malaria parasites. Nature Reviews Microbiology. 2015;13:160–172. doi: 10.1038/nrmicro3402. [DOI] [PubMed] [Google Scholar]
- Dvorin JD, Martyn DC, Patel SD, Grimley JS, Collins CR, Hopp CS, Bright AT, Westenberger S, Winzeler E, Blackman MJ, et al. A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science. 2010;328:910–912. doi: 10.1126/science.1188191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Molecular Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Essien K, Stoeckert CJ., Jr Conservation and divergence of known apicomplexan transcriptional regulons. BMC Genomics. 2010;11:147. doi: 10.1186/1471-2164-11-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naude B, Deitsch KW, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Molecular Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidock DA, Wellems TE. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci U S A. 1997;94:10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flueck C, Bartfai R, Niederwieser I, Witmer K, Alako BT, Moes S, Bozdech Z, Jenoe P, Stunnenberg HG, Voss TS. A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology. PLoS Pathogens. 2010;6:e1000784. doi: 10.1371/journal.ppat.1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flueck C, Bartfai R, Volz J, Niederwieser I, Salcedo-Amaya AM, Alako BT, Ehlgen F, Ralph SA, Cowman AF, Bozdech Z, et al. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathogens. 2009;5:e1000569. doi: 10.1371/journal.ppat.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenal K, Polonais V, Marq JB, Stratmann R, Limenitakis J, Soldati-Favre D. Functional dissection of the apicomplexan glideosome molecular architecture. Cell Host Microbe. 2010;8:343–357. doi: 10.1016/j.chom.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Gao X, Gunalan K, Yap SS, Preiser PR. Triggers of key calcium signals during erythrocyte invasion by Plasmodium falciparum. Nature Communications. 2013;4:2862. doi: 10.1038/ncomms3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nature Biotechnology. 2014;32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco TM, Miteva Y, Conlon FL, Cristea IM. Complementary proteomic analysis of protein complexes. Methods in Molecular Biology. 2012;917:391–407. doi: 10.1007/978-1-61779-992-1_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biology. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han CL, Chien CW, Chen WC, Chen YR, Wu CP, Li H, Chen YJ. A multiplexed quantitative strategy for membrane proteomics: opportunities for mining therapeutic targets for autosomal dominant polycystic kidney disease. Molecular & Cellular Proteomics. 2008;7:1983–1997. doi: 10.1074/mcp.M800068-MCP200. [DOI] [PubMed] [Google Scholar]
- Harding CR, Egarter S, Gow M, Jimenez-Ruiz E, Ferguson DJ, Meissner M. Gliding Associated Proteins Play Essential Roles during the Formation of the Inner Membrane Complex of Toxoplasma gondii. PLoS Pathogens. 2016;12:e1005403. doi: 10.1371/journal.ppat.1005403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EY, Ponts N, Le Roch KG, Lonardi S. Chromatin-driven de novo discovery of DNA binding motifs in the human malaria parasite. BMC Genomics. 2011;12:601. doi: 10.1186/1471-2164-12-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaslip AT, Dzierszinski F, Stein B, Hu K. TgMORN1 is a key organizer for the basal complex of Toxoplasma gondii. PLoS Pathogens. 2010;6:e1000754. doi: 10.1371/journal.ppat.1000754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrocks P, Wong E, Russell K, Emes RD. Control of gene expression in Plasmodium falciparum - ten years on. Molecular & Biochemical Parasitology. 2009;164:9–25. doi: 10.1016/j.molbiopara.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Hu G, Cabrera A, Kono M, Mok S, Chaal BK, Haase S, Engelberg K, Cheemadan S, Spielmann T, Preiser PR, et al. Transcriptional profiling of growth perturbations of the human malaria parasite Plasmodium falciparum. Nature Biotechnology. 2010;28:91–98. doi: 10.1038/nbt.1597. [DOI] [PubMed] [Google Scholar]
- Hume MA, Barrera LA, Gisselbrecht SS, Bulyk ML. UniPROBE, update 2015: new tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Research. 2015;43:D117–122. doi: 10.1093/nar/gku1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iengar P, Joshi NV. Identification of putative regulatory motifs in the upstream regions of co-expressed functional groups of genes in Plasmodium falciparum. BMC Genomics. 2009;10:18. doi: 10.1186/1471-2164-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D, Schureck MA, Desai SA. An essential dual-function complex mediates erythrocyte invasion and channel-mediated nutrient uptake in malaria parasites. eLife. 2017;6 doi: 10.7554/eLife.23485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga S, Kaneko I, Kato T, Yuda M. Identification of an AP2-family Protein That Is Critical for Malaria Liver Stage Development. PLoS One. 2012;7:e47557. doi: 10.1371/journal.pone.0047557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji DD, Arnot DE. A Plasmodium falciparum homologue of the ATPase subunit of a multi-protein complex involved in chromatin remodelling for transcription. Molecular & Biochememical Parasitology. 1997;88:151–162. doi: 10.1016/s0166-6851(97)00089-3. [DOI] [PubMed] [Google Scholar]
- Joshi P, Greco TM, Guise AJ, Luo Y, Yu F, Nesvizhskii AI, Cristea IM. The functional interactome landscape of the human histone deacetylase family. Molecular Systems Biology. 2013;9:672. doi: 10.1038/msb.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josling GA, Petter M, Oehring SC, Gupta AP, Dietz O, Wilson DW, Schubert T, Langst G, Gilson PR, Crabb BS, et al. A Plasmodium falciparum Bromodomain Protein Regulates Invasion Gene Expression. Cell Host Microbe. 2015;17:741–751. doi: 10.1016/j.chom.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Kadekoppala M, Kline K, Akompong T, Haldar K. Stable expression of a new chimeric fluorescent reporter in the human malaria parasite Plasmodium falciparum. Infection & Immunology. 2000;68:2328–2332. doi: 10.1128/iai.68.4.2328-2332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafsack BF, Rovira-Graells N, Clark TG, Bancells C, Crowley VM, Campino SG, Williams AE, Drought LG, Kwiatkowski DP, Baker DA, et al. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature. 2014;507:248–252. doi: 10.1038/nature12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roch KG, Johnson JR, Florens L, Zhou Y, Santrosyan A, Grainger M, Yan SF, Williamson KC, Holder AA, Carucci DJ, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Research. 2004;14:2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Research. 2012;40:D302–305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013. arXiv:13033997v2 [q-bioGN] [Google Scholar]
- LI H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner SE, Swearingen KE, Harupa A, Vaughan AM, Sinnis P, Moritz RL, Kappe SH. Total and putative surface proteomics of malaria parasite salivary gland sporozoites. Molecular & Cellular Proteomics. 2013;12:1127–1143. doi: 10.1074/mcp.M112.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Barragan MJ, Lemieux J, Quinones M, Williamson KC, Molina-Cruz A, Cui K, Barillas-Mury C, Zhao K, Su XZ. Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics. 2011;12:587. doi: 10.1186/1471-2164-12-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rubio JJ, Siegel TN, Scherf A. Malaria: Methods and Protocols. 2012. Genome-wide chromatin immunoprecipitation-sequencing in Plasmodium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorestani A, Sheiner L, Yang K, Robertson SD, Sahoo N, Brooks CF, Ferguson DJ, Striepen B, Gubbels MJ. A Toxoplasma MORN1 null mutant undergoes repeated divisions but is defective in basal assembly, apicoplast division and cytokinesis. PLoS One. 2010;5:e12302. doi: 10.1371/journal.pone.0012302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrzynska K, Pfander C, Chappell L, Yu L, Suarez C, Dundas K, Gomes AR, Goulding D, Rayner JC, Choudhary J, et al. A Knockout Screen of ApiAP2 Genes Reveals Networks of Interacting Transcriptional Regulators Controlling the Plasmodium Life Cycle. Cell Host Microbe. 2017;21:11–22. doi: 10.1016/j.chom.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics. 2009;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehring SC, Woodcroft BJ, Moes S, Wetzel J, Dietz O, Pulfer A, Dekiwadia C, Maeser P, Flueck C, Witmer K, et al. Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum. Genome Biology. 2012;13:R108. doi: 10.1186/gb-2012-13-11-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachebat JA, Ling IT, Grainger M, Trucco C, Howell S, Fernandez-Reyes D, Gunaratne R, Holder AA. The 22 kDa component of the protein complex on the surface of Plasmodium falciparum merozoites is derived from a larger precursor, merozoite surface protein 7. Molecular & Biochemical Parasitology. 2001;117:83–89. doi: 10.1016/s0166-6851(01)00336-x. [DOI] [PubMed] [Google Scholar]
- Painter HJ, Altenhofen LM, Kafsack BF, Llinas M. Whole-genome analysis of Plasmodium spp Utilizing a new agilent technologies DNA microarray platform. Methods in Molecular Biology. 2013;923:213–219. doi: 10.1007/978-1-62703-026-7_14. [DOI] [PubMed] [Google Scholar]
- Painter HJ, Campbell TL, Llinas M. The Apicomplexan AP2 family: integral factors regulating Plasmodium development. Molecular & Biochemical Parasitology. 2011;176:1–7. doi: 10.1016/j.molbiopara.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proellocks NI, Coppel RL, Waller KL. Dissecting the apicomplexan rhoptry neck proteins. Trends in Parasitology. 2010;26:297–304. doi: 10.1016/j.pt.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Research. 2014;42:W187–191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly HB, Wang H, Steuter JA, Marx AM, Ferdig MT. Quantitative dissection of clone-specific growth rates in cultured malaria parasites. International Journal of Parasitology. 2007;37:1599–1607. doi: 10.1016/j.ijpara.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard D, Kats LM, Langer C, Black CG, Mitri K, Boddey JA, Cowman AF, Coppel RL. Identification of rhoptry trafficking determinants and evidence for a novel sorting mechanism in the malaria parasite Plasmodium falciparum. PLoS Pathogens. 2009;5:e1000328. doi: 10.1371/journal.ppat.1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell K, Emes R, Horrocks P. Triaging informative cis-regulatory elements for the combinatorial control of temporal gene expression during Plasmodium falciparum intraerythrocytic development. Parasites & Vectors. 2015;8:81. doi: 10.1186/s13071-015-0701-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo I, Oksman A, Goldberg DE. Fatty acid acylation regulates trafficking of the unusual Plasmodium falciparum calpain to the nucleolus. Molecular Microbiol. 2009;72:229–245. doi: 10.1111/j.1365-2958.2009.06639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PR, Kats LM, Drew DR, O'Donnell RA, O'Neill M, Maier AG, Coppel RL, Crabb BS. A set of glycosylphosphatidyl inositol-anchored membrane proteins of Plasmodium falciparum is refractory to genetic deletion. Infection & Immunology. 2006;74:4330–4338. doi: 10.1128/IAI.00054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangurdekar DP. R package version 1.4.0. Bioconductor. 2014. Rnits: R Normalization and Inference of Time Series data. [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherling ES, Knuepfer E, Brzostowski JA, Miller LH, Blackman MJ, Ooij CV. The Plasmodium falciparum rhoptry protein RhopH3 plays essential roles in host cell invasion and nutrient uptake. eLife. 2017;6 doi: 10.7554/eLife.23239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Molecular Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Chitnis CE. Signalling mechanisms involved in apical organelle discharge during host cell invasion by apicomplexan parasites. Microbes and Infection. 2012;14:820–824. doi: 10.1016/j.micinf.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Sinha A, Hughes KR, Modrzynska KK, Otto TD, Pfander C, Dickens NJ, Religa AA, Bushell E, Graham AL, Cameron R, et al. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature. 2014;507:253–257. doi: 10.1038/nature12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann T, Fergusen DJ, Beck HP. etramps a new Plasmodium falciparum gene family coding for developmentally regulated and highly charged membrane proteins located at the parasite-host cell interface. Molecular Biology of the Cell. 2003;14:1529–1544. doi: 10.1091/mbc.E02-04-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spycher C, Rug M, Pachlatko E, Hanssen E, Ferguson D, Cowman AF, Tilley L, Beck HP. The Maurer's cleft protein MAHRP1 is essential for trafficking of PfEMP1 to the surface of Plasmodium falciparum -infected erythrocytes. Molecular Microbiology. 2008;68:1300–1314. doi: 10.1111/j.1365-2958.2008.06235.x. [DOI] [PubMed] [Google Scholar]
- Striepen B, Jordan CN, Reiff S, van Dooren GG. Building the perfect parasite: cell division in apicomplexa. PLoS Pathogens. 2007;3:e78. doi: 10.1371/journal.ppat.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova ES, Francia M, Striepen B, White MW. A novel bipartite centrosome coordinates the apicomplexan cell cycle. PLoS Biology. 2015;13:e1002093. doi: 10.1371/journal.pbio.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton TJ, Iyer LM, Anantharaman V, Enomoto S, Abrahante JE, Subramanian GM, Hoffman SL, Abrahamsen MS, Aravind L. Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Research. 2004;14:1686–1695. doi: 10.1101/gr.2615304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham WH, Healer J, Cowman AF. Erythrocyte and reticulocyte binding-like proteins of Plasmodium falciparum. Trends in Parasitology. 2012;28:23–30. doi: 10.1016/j.pt.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Trucco C, Fernandez-Reyes D, Howell S, Stafford WH, Scott-Finnigan TJ, Grainger M, Ogun SA, Taylor WR, Holder AA. The merozoite surface protein 6 gene codes for a 36 kDa protein associated with the Plasmodium falciparum merozoite surface protein-1 complex. Molecular & Biochemical Parasitology. 2001;112:91–101. doi: 10.1016/s0166-6851(00)00350-9. [DOI] [PubMed] [Google Scholar]
- Volz J, Carvalho TG, Ralph SA, Gilson P, Thompson J, Tonkin CJ, Langer C, Crabb BS, Cowman AF. Potential epigenetic regulatory proteins localise to distinct nuclear sub-compartments in Plasmodium falciparum. International Journal of Parasitology. 2010;40:109–121. doi: 10.1016/j.ijpara.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Walker DM, Oghumu S, Gupta G, McGwire BS, Drew ME, Satoskar AR. Mechanisms of cellular invasion by intracellular parasites. Cellular and Molecular Life Sciences. 2014;71:1245–1263. doi: 10.1007/s00018-013-1491-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss GE, Gilson PR, Taechalertpaisarn T, Tham WH, de Jong NW, Harvey KL, Fowkes FJ, Barlow PN, Rayner JC, Wright GJ, et al. Revealing the Sequence and Resulting Cellular Morphology of Receptor-Ligand Interactions during Plasmodium falciparum Invasion of Erythrocytes. PLoS Pathogens. 2015;11:e1004670. doi: 10.1371/journal.ppat.1004670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Mann M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. Journal of Proteome Research. 2009;8:5674–5678. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- Wright GJ, Rayner JC. Plasmodium falciparum erythrocyte invasion: combining function with immune evasion. PLoS Pathogens. 2014;10:e1003943. doi: 10.1371/journal.ppat.1003943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Johnson JR, Benner C, Yan SF, Chen K, Le Roch KG, Zhou Y, Winzeler EA. In silico discovery of transcription regulatory elements in Plasmodium falciparum. BMC Genomics. 2008;9:70. doi: 10.1186/1471-2164-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Huang Y, Zhang Y, Fang X, Claes A, Duchateau M, Namane A, Lopez-Rubio JJ, Pan W, Scherf A. A critical role of perinuclear filamentous actin in spatial repositioning and mutually exclusive expression of virulence genes in malaria parasites. Cell Host Microbe. 2011;10:451–463. doi: 10.1016/j.chom.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.