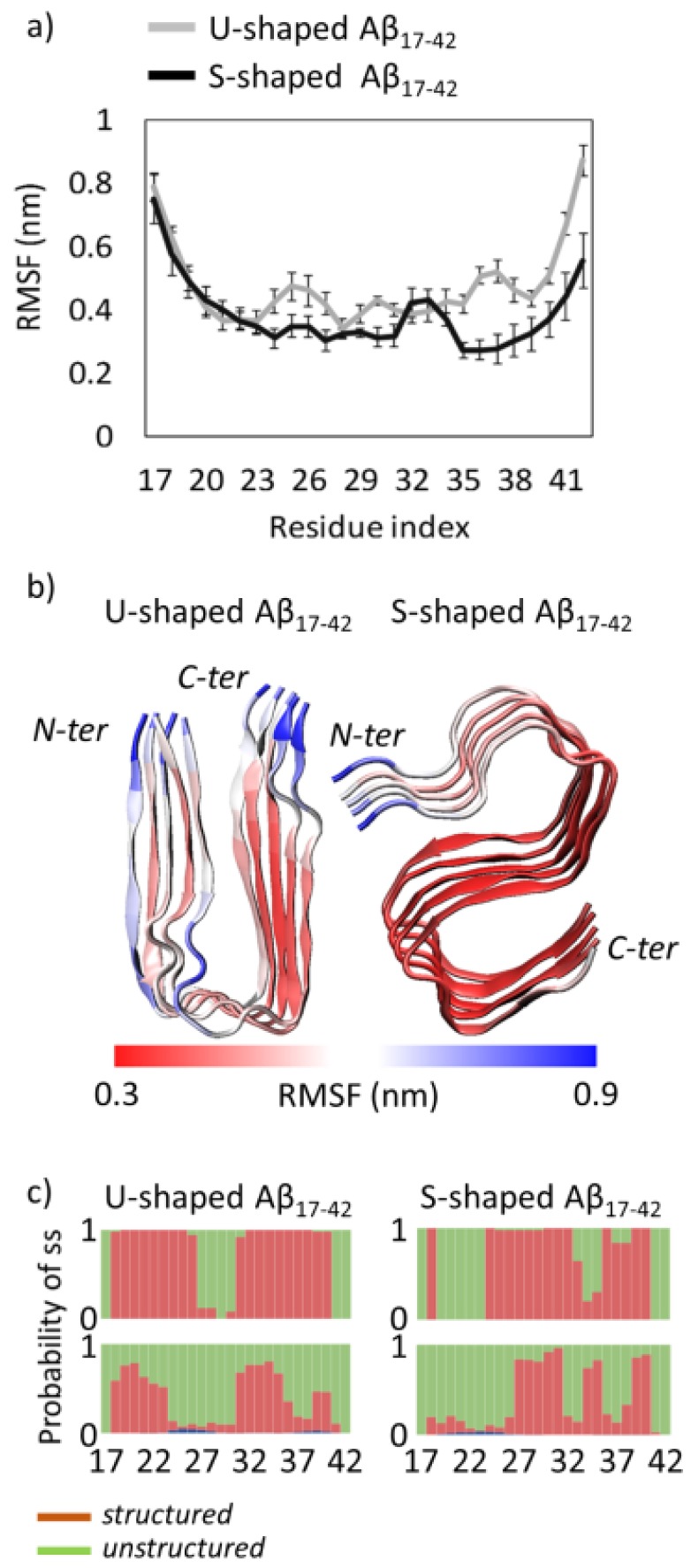
Figure 1.
(a) U-shaped and S-shaped Root Mean Square Fluctuation (RMSF) of atomic positions averaged on each protein residue. Each average value and relative standard deviation was obtained by mediating the RMSF on the five considered protein chains (A–E); (b) U-shaped and S-shaped structural models coloured on the basis of RMSF values. The scale bar moves from red (RMSF = 0.3 nm) to blue (RMSF = 0.9 nm); (c) U-shaped and S-shaped residue secondary structure probability, calculated over 5 considered chains (A–E) in the PDB models (upper row) and on the Replica Exchange Molecular Dynamics (REMD) ensemble at 300 K (lower row). For the sake of clarity, the secondary structures are classified in structured (red) and unstructured (green). Moreover, the structured class does not contain helices (shown in blue) being their contribution negligible throughout the overall REMD ensemble at 300 K.