Abstract
Agrobacterium tumefaciens can genetically transform various eukaryotic cells because of the presence of a resident tumor-inducing (Ti) plasmid. During infection, a defined region of the Ti plasmid, transfer DNA (T-DNA), is transferred from bacteria into plant cells and causes plant cells to abnormally synthesize auxin and cytokinin, which results in crown gall disease. T-DNA and several virulence (Vir) proteins are secreted through a type IV secretion system (T4SS) composed of T-pilus and a transmembrane protein complex. Three members of Arabidopsis reticulon-like B (RTNLB) proteins, RTNLB1, 2, and 4, interact with VirB2, the major component of T-pilus. Here, we have identified that other RTNLB proteins, RTNLB3 and 8, interact with VirB2 in vitro. Root-based A. tumefaciens transformation assays with Arabidopsis rtnlb3, or rtnlb5-10 single mutants showed that the rtnlb8 mutant was resistant to A. tumefaciens infection. In addition, rtnlb3 and rtnlb8 mutants showed reduced transient transformation efficiency in seedlings. RTNLB3- or 8 overexpression transgenic plants showed increased susceptibility to A. tumefaciens and Pseudomonas syringae infection. RTNLB1-4 and 8 transcript levels differed in roots, rosette leaves, cauline leaves, inflorescence, flowers, and siliques of wild-type plants. Taken together, RTNLB3 and 8 may participate in A. tumefaciens infection but may have different roles in plants.
Keywords: RTNLB, Agrobacterium
1. Introduction
In nature, the phytopathogenic bacterium Agrobacterium tumefaciens of the family Rhizobiaceae infects susceptible plants and causes crown gall tumors. The disease results from the transfer of effector virulence (Vir) proteins and the transfer DNA (T-DNA) derived from a large bacterial tumor-inducing (Ti) plasmid. T-DNA transfer from A. tumefaciens into a plant cell requires the expression of several virulence (vir) genes that reside on the Ti plasmid [1,2,3,4]. The uncontrolled growth of crown gall tumors results from the transfer and expression of oncogenes encoded by the wild-type T-DNA, which directs overproduction of the plant growth hormones cytokinin and auxin [5]. Another set of genes in wild-type T-DNA causes the production of bacterial nutrients, called opines, which are then utilized by A. tumefaciens as a carbon and sometimes nitrogen source.
A. tumefaciens uses a VirA/VirG two-component regulatory system to sense various environmental signals, including acidity, monosaccharides, and phenolic compounds, and induce vir gene expression [6,7]. With the help of VirD1 and VirD2 proteins, the single-stranded T-DNA is processed and then transported into plants via a type IV secretion system (T4SS). The T4SS is used by many pathogens to deliver protein and/or DNA into the cell cytosol and modulate eukaryotic cell functions [8,9,10,11]. The process involves the recognition of cognate substrates and delivery of the substrates across membrane barrier(s).
The T4SS consists of two functional components, a transmembrane transporter comprising VirD4 and VirB1-11 proteins, and a filamentous pilus (T-pilus) [12,13,14]. The T-pilus is a long, semi-rigid, flexuous filament 10 nm in diameter that may play an important role in virulence. The T-pilus contains at least two VirB proteins. The major component, VirB2, is translated as a 12.3-kD pro-pilin protein but is processed to a 7.2-kD pilin protein by removal of a N-terminal signal peptide (1–47 amino acid residues) [15,16,17]. T-pilin, 74 amino acid residues long, is coupled between the amino terminal residue Gln-48 to Gly-121 at the carboxy terminus in a head-to-tail peptide bond, forming an unusual cyclic peptide [18]. VirB5 co-fractionates as a minor component in T-pilus preparations and contributes to T-pilus assembly [19]. VirB5 is localized at the tips of the cell-bound T-pili and might mediate host cells and bacteria contact via interactions with the host protein during A. tumefaciens infection [20]. After T-DNA enters plant cells, T-DNA, along with the attached VirD2 protein, will be transported into the plant nucleus and integrated into the plant chromosome with the assistance of VirE2, VirF, other Vir proteins, and plant proteins. During T-DNA nuclear import, VirE2 may interact with the plant VirE2-interacing plant protein (VIP1) in the cytoplasm to assist in nuclear targeting of T-DNA and to block endogenous VIP1 from activating plant defense responses [4,21,22]. Successful A. tumefaciens-mediated plant transformation involves a continuous battle of plant cells activating a defense response to repel bacterial infection and bacteria using Vir proteins and manipulating plant proteins to elude the plant’s immunity systems.
A previous study [23] identified plant-encoded proteins that may mediate the initial contact of A. tumefaciens T-pilus with the host cell. Yeast two-hybrid and in vitro assays revealed two classes of Arabidopsis proteins that interact with VirB2. The first class consists of three related proteins: reticulon-like protein B1 (RTNLB1), 2, and 4. The second class is a RAB8B GTPase. Yeast two-hybrid assay and in vitro interaction studies demonstrated that the three RTNLB proteins interact with themselves, each other, and RAB8B, so these proteins may form a multimeric complex [23]. Pre-incubation of induced A. tumefaciens with GST-RTNLB1 protein reduced the A. tumefaciens transformation efficiency of Arabidopsis suspension cells. The level of RTNLB1 protein transiently increased immediately after A. tumefaciens infection. Arabidopsis rtnlb1 mutant plants were recalcitrant to Agrobacterium-mediated transformation, whereas Arabidopsis RTNLB1-overexpressing transgenic plants were hypersusceptible to A. tumefaciens infection [23]. The three RTNLB proteins all have a carboxyl-terminal 150–201 amino acid reticulon (RTN) homology domain composed of two large hydrophobic regions and a ~66 amino acid loop in between. The RTN1 protein, a membrane-anchored component of the endoplasmic reticulum (ER), is the first identified member of this family and is expressed in the central nervous system and in neuroendocrine cells [24,25,26]. RTN proteins may interact with themselves or recruit other proteins to form a complex and perform specific functions. In mammalian, yeast, and plant cells, RTN proteins are involved in various endomembrane-related processes, which includes intracellular transport, vesicle formation, and membrane curvature [27,28,29,30,31,32,33].
More than 250 reticulon-like (RTNL) genes have been identified in divergent eukaryotes, fungi, plants, and animals. RTNL genes appear to have evolved from an intron-rich ancestor [27,34]. There are 21 RTNLB proteins in Arabidopsis thaliana sharing amino acid sequence similarity to the reticulon domain at the C terminus [30,35]. Consistent with the peripheral location of RTNLB1-GFP [23], RTNLB1 and RTNLB6 were found by proteomic analyses of plasma membrane-enriched preparations [36]. Fluorescent-labeled RTNLB2, 4, [35] and RTNLB13 [31] are localized in ER tubules. RTNLB1-4 and 13 can co-localize and constrict tubular ER membranes, so RTNLB proteins may bend the membrane and form multimeric, arc-like structures to shape the ER tubules [33]. In addition, the C-terminal RHD domain is required for RTNLB1-4 to reside in ER membranes and efficiently constrict ER tubules but is not necessary for their homo- and heterotypic interactions [33].
The RTNLB3 and 6 proteins may participate the formation of the desmotubule, membrane structures derived from the cortical ER that transverse through plasmodesmata (PD) [37]. Many viral movement proteins can help viruses spread via interactions with the PD [37,38,39]. RTNLB3 and 6 co-localize with the viral movement protein of Tobacco mosaic virus at the primary PD [37]. Potato virus X movement protein is also detected in the desmotubules of Nicotiana benthamiana PD [39]. A protein microarray screen identified RTNLB1 and 2 proteins that interact with the Arabidopsis FLAGELIN-SENSITIVE2 (FLS2) protein, one of the pattern recognition receptors (PRRs) for the bacterial flagellin [40]. The rtnlb1,2 double mutant and RTNLB1 overexpression plants show increased susceptibility to Pseudomonas syringae pv. tomato DC3000 (Pst) infection and decreased FLS2-mediated immunity responses [40]. FLS2 levels at the plasma membrane are lower in the rtnlb1,2 double mutant and RTNLB1 overexpression plants, so RTNLB1 and 2 may control the trafficking of the FLS2 protein to the plasma membrane [40]. However, relatively little is known about the function of RTNLB proteins in plant–microbe interactions.
In this study, we further identified two additional RTNLB proteins, RTNLB3 and 8, that interact with the A. tumefaciens VirB2 protein. A. tumefaciens-mediated transient transformation efficiency was lower in rtnlb3 and rtnlb8 mutant than wild-type plants. Furthermore, overexpression of RTNLB3 or 8 in transgenic Arabidopsis plants enhanced both stable and transient A. tumefaciens transformation efficiency. Also, RTNLB3 or 8 overexpression plants were hypersusceptible to Pst DC3000 infection. This study further reveals the involvement of RTNLB3 and 8 in plant–microbe interactions.
2. Results
2.1. Interactions Among RTNLB3 and 8 and Vir Proteins in Yeast and In Vitro
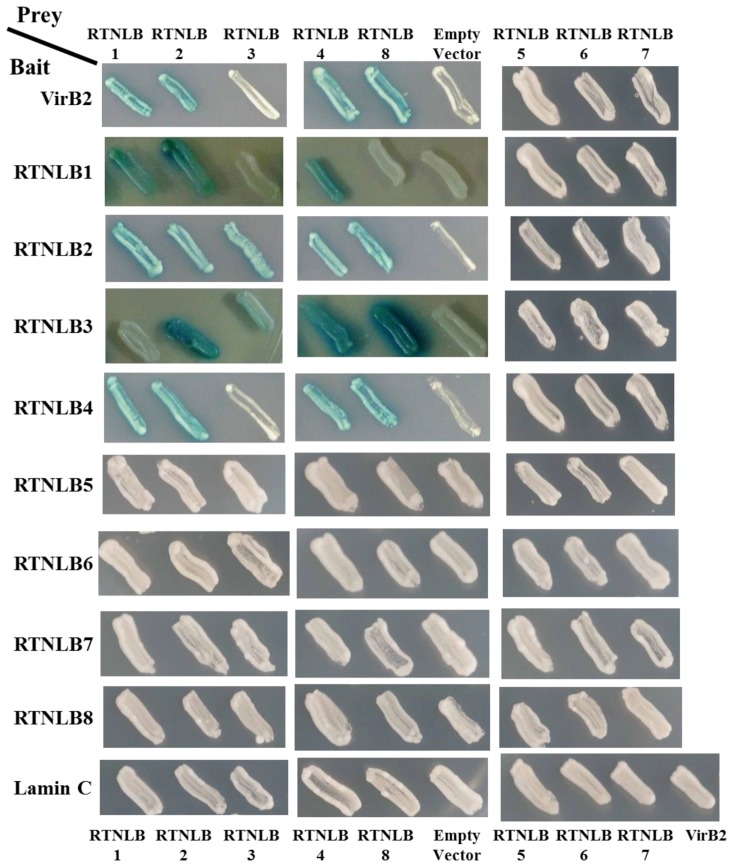
A previous study demonstrated that RTNLB1, 2, and 4 interacted with the C-terminal-processed portion of VirB2 protein in yeast two-hybrid and in vitro assays [23]. From the phylogenetic tree results of the Arabidopsis RTNLB family, RTNLB1-8 proteins belong to the Group I proteins containing an N-terminal domain with 43–93 amino acid residues and a short C-terminal domain [27,30]. Therefore, we cloned RTNLB3 and RTNLB5-8 from Arabidopsis cDNA and examined whether RTNLB3 and RTNLB5-8 could interact with A. tumefaciens VirB2 bait protein in yeast two-hybrid assays. The RTNLB8 prey protein but not the RTNLB3 and RTNLB5-7 proteins interacted with the VirB2 bait protein in yeast (Figure 1). RTNLB1, 2, and 4 proteins interacted with the VirB2 protein as well, which was consistent with previous results [23], and were used as positive controls in the yeast two-hybrid assays. As expected, the RTNLB1-8 prey proteins did not interact with the unrelated Lamin C bait protein in yeast and was used as the negative control (Figure 1). We also examined whether RTNLB3 and RTNLB5-8 proteins could interact with other Vir proteins, including VirB5 (the minor component of T-pili), VirB1, ViB1*, VirD2, VirE1, VirE2, and VirF. RTNLB3 and RTNLB5-8 did not interact with other tested Vir proteins, which was similar to the results for RTNLB1, 2, and 4 proteins [23].
Figure 1.
RTNLB8, not 3, and 5–7 proteins, interacted with the processed VirB2 in yeast. RTNLB1-8 proteins were tested for interactions with VirB2 or the RTNLB1-8 using a yeast two-hybrid assay. The RTNLB5-7 proteins showed no interactions with RTNLB1-8 proteins in yeast. The unrelated Lamin C bait protein was the negative control.
Previous studies have demonstrated that RTNLB1, 2, and 4 can interact with each other and with themselves [23,33]. We next tested whether the RTNLB3 and RTNLB5–8 proteins interacted with themselves and/or other RTNLB1-8 proteins in yeast two-hybrid assays. RTNLB2 used as a bait fusion protein interacted with the RTNLB3 or 8 but not RTNLB5, 6, or 7 (Figure 1 and Table S1). RTNLB3 used as the bait fusion protein interacted with RTNLB2, 4 or 8 but not RTNLB3 or RTNLB5-7. Figure 1 results demonstrated that the RTNLB4 protein interacted with RTNLB8 but not RTNLB3 or RTNLB5-7. As well, RTNLB5, 6, 7, or 8 used as bait fusion proteins did not interact with RTNLB1-8 proteins in yeast (Figure 1). Similarly, RTNLB1 did not interact with the RTNLB3 or RTNLB5-8 proteins in yeast two-hybrid assays (Figure 1). Some of the positive yeast two-hybrid interactions were not observed when the tested bait protein was swapped with the prey proteins (Figure 1 and Table S1). For example, the RTNLB2 bait protein interacted with the RTNLB8 prey protein in yeast; whereas the RTNLB8 bait protein did not interact with the RTNLB2 prey protein. These inconsistent findings may result from different conformations of the bait and prey fusion proteins in yeast, as previously reported for interactions between RTNLB and RAB8 [23].
We next performed β-galactosidase activity assays to quantify the interaction strengths in these yeast strains. The white colony yeast strains on the SD media with X-gal substrates showed zero β-galactosidase activity, so the liquid-based β-galactosidase activity assays showed similar results as the plate-based yeast two-hybrid assays (Table S1). Yeast strains expressing the RTNLB3 bait with RTNLB2, 4, or 8 prey proteins showed relatively lower β-galactosidase activities as compared with yeast strains expressing the RTNLB2 or 4 bait proteins with the same tested prey proteins, which suggests that the interaction strengths might be lower among RTNLB3 interacting with other RTNLB proteins (Table S1).
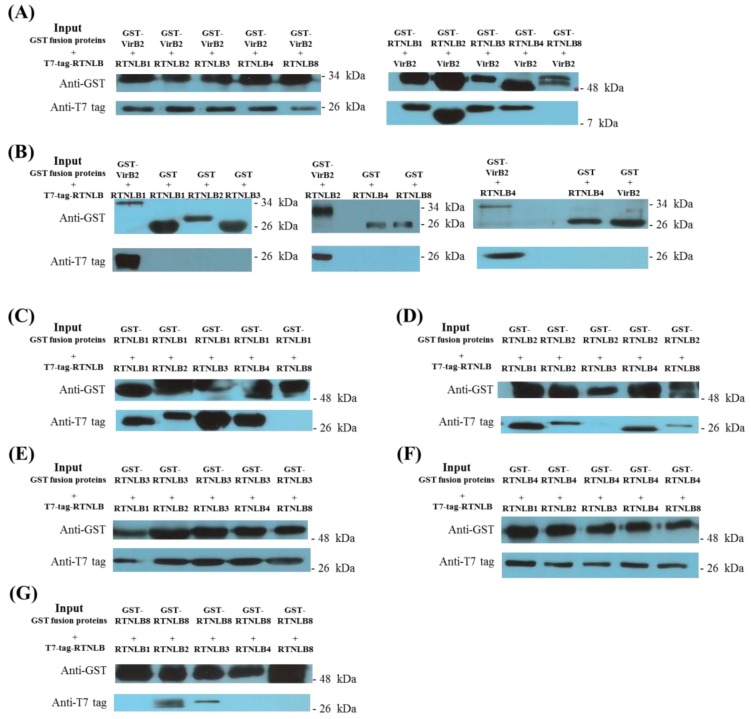
In vitro glutathione-S-transferase (GST) pull-down assays were used to determine direct protein–protein interactions of RTNLB3 and 8 with VirB2 and other RTNLB proteins. T7-tagged-RTNLB1, 2, 3, 4, and 8 proteins interacted with the GST-VirB2 fusion protein but not the GST protein in vitro (Figure 2A,B). In addition, GST-RTNLB1, 2, 3, and 4 fusion proteins but not the GST-RTNLB8 fusion protein interacted with the T7-tagged-VirB2 protein. The in vitro protein interactions between RTNLB1-4 and RTNLB8 were examined with GST pull-down assays by using the GST fusions and T7-tagged versions of RTNLB1-4 and 8. GST-RTNLB1 fusion proteins interacted with T7-tagged-RTNLB1, 2, 3, 4, but not T7-tagged-RTNLB8 proteins in vitro, whereas GST-RTNLB2 fusion protein interacted with T7-tagged-RTNLB1, 2, 4, 8, but not T7-tagged-RTNLB3 protein in vitro (Figure 2C,D). Furthermore, GST-RTNLB3 and GST-RTNLB4 fusion proteins interacted with the five tested RTNLB proteins (Figure 2E,F). However, only T7-tagged-RTNLB2 and 3 directly interacted with the GST-RTNLB8 fusion protein (Figure 2G). These interaction results were consistent with previous observations showing that RTNLB1-4 proteins may have homo- and heterotypic interactions [33]. The yeast two-hybrid assay and GST pull-down assay results summarized in Table S1 indicate that RTNLB3 interacted with VirB2, RTNLB1-4 and 8; whereas RTNLB8 interacted with VirB2, and RTNLB2, 3, 4. Interestingly, more positive interaction results of RTNLB3 and 8 with VirB2 and with other RTNLB proteins were obtained with GST pull-down than yeast two-hybrid assays. Previous studies in plants have suggested that RTNLB proteins are membrane proteins and are mainly localized in the plant endomembrane systems [31,33,35,36]. Therefore, the various fusion versions of RTNLB proteins used in our yeast two-hybrid and GST pull-down assays may not form the same conformation as native RTNLB protein in plant cells.
Figure 2.
The GST-VirB2 fusion protein interacted with RTNLB1-4 and 8 proteins in vitro. The GST-fusion and GST only proteins were linked with glutathione-sepharose beads and incubated with T7-tagged proteins to test their interactions in vitro. Bound proteins were eluted with glutathione and analyzed by protein gel blot using anti-T7 tag and anti-GST antibodies. Panel A, interactions between VirB2 and RTNLB1-4 and 8 were determined by using the GST-VirB2 fusion protein and T7-tagged-RTNLB1-4 and 8 or the GST-fusion of RTNLB1-4 and 8 and the T7-tagged-VirB2 protein. Panel B, the GST-only protein was used as a negative control in GST pull-down assays. The GST fusions of RTNLB1 (Panel C), RTNLB2 (Panel D), RTNLB3 (Panel E), RTNLB4 (Panel F), and RTNLB8 (Panel G) were used to investigate their interactions with T7-tagged-RTNLB1-4 and 8 proteins.
2.2. Arabidopsis Rtnlb3 and Rtnlb8 Mutants Showed Reduced Levels of A. tumefaciens-Mediated Transformation Efficiency
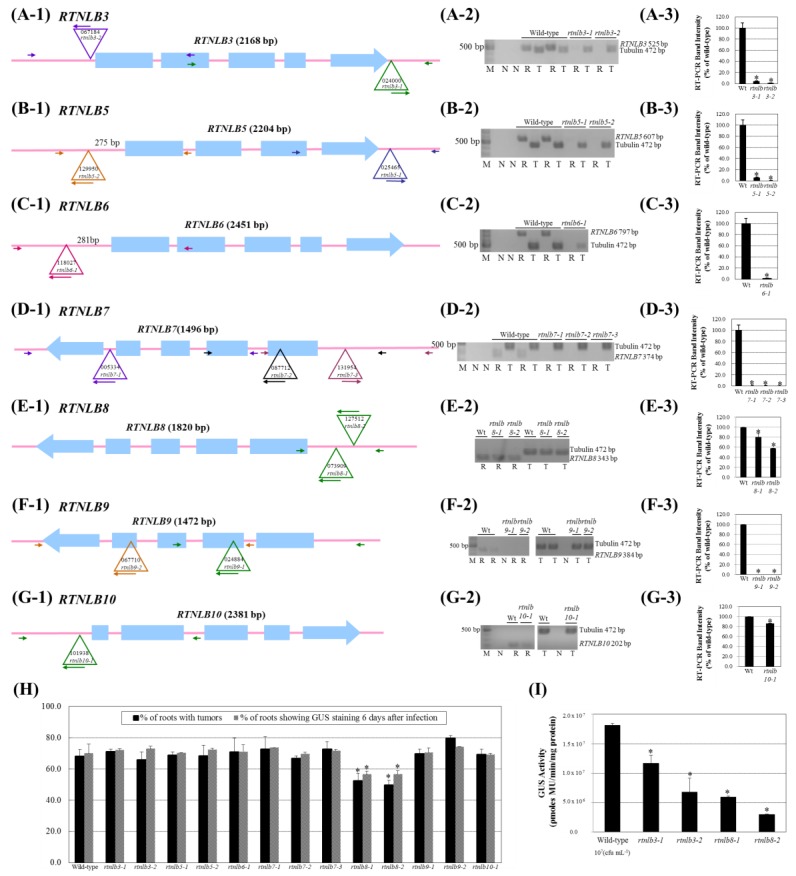
Because the RTNLB3 and 8 proteins interacted with the A. tumefaciens VirB2 protein in vitro, we next examined whether the RTNLB3 and RTNLB5-10 proteins are involved in the A. tumefaciens infection process. We obtained several T-DNA insertions Arabidopsis mutants of RTNLB3 or RTNLB5-10 genes (Table S2) and tested susceptibilities of various rtnlb mutant plants to root- and seedling-based A. tumefaciens infection assays. At least one T-DNA insertion homozygous mutant was identified for RTNLB3 and RTNLB5-10 (Table S2 and Figure 3(A-1–A-7)). In the rtnlb3, rtnlb5, rtnlb6, rtnlb8, and rtnlb10 single mutants, the T-DNA insertion sites were mainly located in the 3′ or 5′ untranslated region (UTR) of RTNLB genes (Table S2 and Figure 3(A-1–A-3,A-5,A-7)). The T-DNA insertion sites of the rtnlb7 and rtnlb9 single mutants were in the intron or exon (Table S2 and Figure 3(A-4,A-6)). Semi-quantitative RT-PCR results showed that RTNLB target gene transcript levels were reduced to less than 5% of the wild-type level or were not detectable in the rtnlb3, rtnlb5, rtnlb6, rtnlb7, and rtnlb9 single mutants (Figure 3(B-1–B-7)) (Figure 3(C-1–C-4,C-6)), which suggests that T-DNA insertions in these mutants may significantly affect the target RTNLB gene transcript stability and accumulation. In the rtnlb8 and rtnlb10 single mutants, the target gene transcript levels decreased to 58% to 86% of the wild-type levels (Figure 3(C-5,C-7)). We next examined transformation frequencies of these rtnlb mutants with stable and transient A. tumefaciens-mediated root transformation. Only rtnlb8-1 and rtnlb8-2 mutants showed lower levels of tumor formation and transient transformation efficiency than wild-type plants (Figure 3D), whereas other tested rtnlb mutants were as susceptible to transformation by A. tumefaciens as wild-type plants. Because transient transformation does not require T-DNA integration into the plant genome [41], these data suggest that the transformation process may be blocked at a step before T-DNA integration in the rtnlb8 mutants and RTNLB8 may be involved at the early step(s) in A. tumefaciens-mediated root transformation process. Different types of plant tissues may show different susceptibility to A. tumefaciens infection [42]. We therefore used Arabidopsis seedlings for transient transformation assays [43] with the rtnlb3 and rtnlb8 mutants. GUS activities were decreased 36% to 63% in the rtnlb3-1 and rtnlb3-2 single mutants and 67% to 84% in the rtnlb8-1 and rtnlb8-2 single mutants as compared with wild-type plants (Figure 3E). The decreased levels of GUS activities were greater in the rtnlb8 than rtnlb3 mutant (Figure 3E), which suggests that low expression of the RTNLB8 gene in the mutant plants might affect the A. tumefaciens-mediated transient transformation efficiency of seedlings more than the RTNLB3 gene. The rtnlb3 mutants showed lower transformation efficiency than wild-type plants with only the seedling-based transformation assay and not the root-based assays (Figure 3D,E), which suggests that seedling tissues might be more sensitive to A. tumefaciens infection than root tissues and/or the RTNLB3 gene might participate in efficient A. tumefaciens infection of Arabidopsis seedlings. In addition, RTNLB5, 6, 7, 9, and 10 might not be directly involved in the A. tumefaciens infection process.
Figure 3.
The Arabidopsis rtnlb3 and rtnlb8 T-DNA insertion mutant seedlings were resistant to A. tumefaciens infection. Panel A, schematic representations of the T-DNA insertion regions around the Arabidopsis RTNLB3 (Panel A-1), RTNLB5 (Panel A-2), RTNLB6 (Panel A-3), RTNLB7 (Panel A-4), RTNLB8 (Panel A-5), RTNLB9 (Panel A-6), and RTNLB10 (Panel A-7) genes. Blue boxes represented exon regions of each RTNLB gene. The large open triangle represents T-DNA insertion sites in each RTNLB gene. The long and short arrows indicate the locations of primers used in genomic DNA PCR analysis. Panel B, RT-PCR results of target RTNLB transcripts in rtnlb3 and rtnlb5-10 single mutants. The α-tubulin was an internal control. Panel C, transcript levels of each RTNLB gene in rtnlb single mutants shown as a relative percentage of wild-type plants. Data are mean ± SE from at least 3 RT-PCR reactions of each mutant. Panel D, transformation efficiencies of rtnlb8-1 and rtnlb8-2 and wild-type plants. Black bars indicate the percentage of root segments forming tumors 1 month after infection with 108 cfu·mL−1 tumorigenic A. tumefaciens A208 strain. Grey bars show the percentage of root segments with GUS activity 6 days after infection with 108 cfu·mL−1 A. tumefaciens At849 strain. Panel E, rtnlb3 and rtnlb8 mutant seedlings showed decreased susceptibility to transient transformation. Transient transformation efficiency in mutant seedlings infected with 107 cfu·mL−1 acetosyringone (AS)-induced A. tumefaciens strain for 3 days. Data are mean ± SE. * p < 0.05 compared with the wild-type by pairwise Student’s t test.
2.3. RTNLB3 and 8 Overexpression Increased Plant Susceptibility to A. tumefaciens-Mediated Transformation
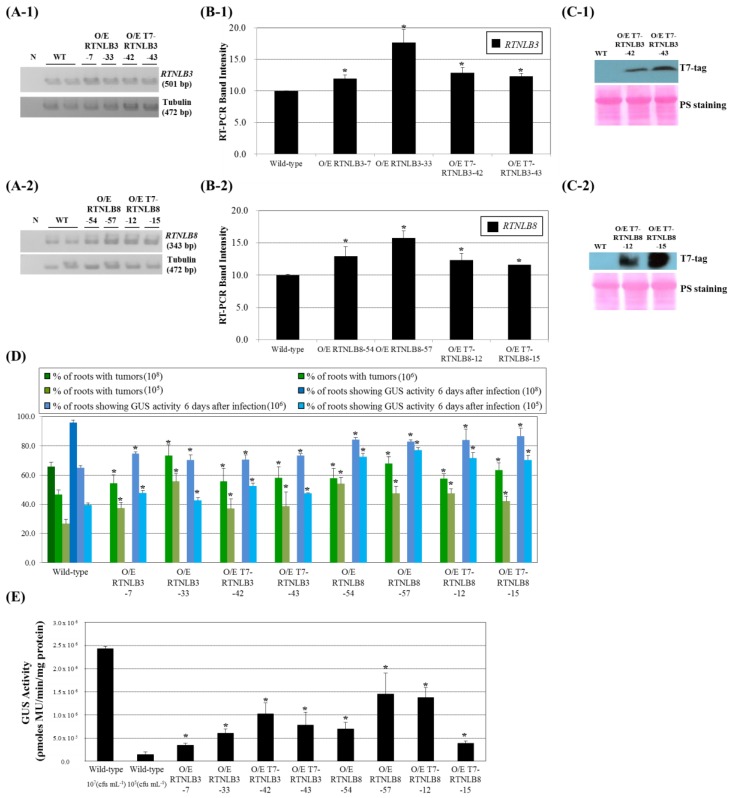
Because the rtnlb3 and rtnlb8 seedling plants were recalcitrant to A. tumefaciens infection, we next determined whether overexpression of RTNLB3 or 8 in plants could enhance the efficiency of A. tumefaciens infection. RTNLB3 and 8 and T7-tagged-RTNLB3 and 8 genes were individually overexpressed in transgenic plants by using a double CaMV 35S promoter. RTNLB3 transcript level was increased 1.2- to 1.7-fold in RTNLB3 and T7-tagged-RTNLB3 overexpression plants (Figure 4(A-1,B-1)). Similarly, RTNLB8 transcript level was increased 1.2- to 1.6-fold in RTNLB8 and T7-tagged-RTNLB8 overexpression plants (Figure 4(A-2,B-2)). Protein gel blot analysis with anti-T7-tag antibody demonstrated that T7-tagged-RTNLB3 or 8 recombinant proteins were highly accumulated in transgenic plants (Figure 4(C-1,C-2)). Root tissues of the RTNLB3 and 8 and T7-tagged-RTNLB3 and 8 overexpression plants were then infected with relatively lower concentrations of A. tumefaciens, 105 and 106 cfu·mL−1. Overexpression of RTNLB3 or T7-tagged-RTNLB3 in transgenic plants increased transient transformation efficiency 1.1- to 1.3-fold and enhanced tumor formation rates 1.2- to 2.1-fold as compared with wild-type plants (Figure 4D). Similarly, both stable and transient transformation rates of RTNLB8 and T7-tagged-RTNLB8 overexpression plants were increased 1.3- to 2.0-fold as compared with wild-type plants (Figure 4D). These data demonstrate that overexpression of RTNLB3 or RTNLB8 in plants enhanced transgenic plant root tissue susceptibility to A. tumefaciens and the presence of the T7 tag sequence in the N-terminal region of RTNLB3 and 8 proteins may not affect the RTNLB protein functions during A. tumefaciens infection. With 105 cfu·mL−1 A. tumefaciens used to infect the RTNLB3 and 8 overexpression seedlings, RTNLB3 overexpression plants showed increased GUS activities by 2.4- to 7.2-fold, whereas RTNLB8 overexpression plants showed increased GUS activities by 2.7- to 10.1-fold as compared with wild-type plants (Figure 4E). Taken together, these data indicated that the RTNLB3 and 8 may play important roles in plants during A. tumefaciens infection.
Figure 4.
RTNLB3 and RTNLB8 overexpression (O/E) transgenic plants were hypersusceptible to A. tumefaciens infections. Panel A, RT-PCR analysis of RTNLB transcript levels in RTNLB3 (Panel A-1) and RTNLB8 (Panel A-2) O/E plants and the wild type. The α-tubulin was used as an internal control. Panel B, transcript levels of RTNLB3 (Panel B-1) or RTNLB8 (Panel B-2) in O/E plants relative to wild-type expression. Data are mean ± SE from at least 3 RT-PCR reactions of each mutant. Panel C, the T7-tagged-RTNLB3 (Panel C-1) and RTNLB8 (Panel C-2) O/E plants accumulated T7-tagged RTNLB proteins. Ponceau S (PS) staining was used to show equivalent loading of total protein in each lane. Panel D, Transient transformation efficiency of RTNLB3 and 8 O/E and wild-type plants. Green bars represent the percentage of root segments developing tumors after infection with 108, 106, or 105 cfu·mL−1 of A. tumefaciens A208. Blue bars indicate the percentage of root segments with GUS activity after infection with 108, 106, or 105 cfu·mL−1 of A. tumefaciens At849 strain. The 108 cfu mL−1 of A. tumefaciens was used to infect wild-type roots as a positive control to indicate successful transformation. Panel E, Enhanced transient transformation efficiency in seedlings of RTNLB3 and 8 O/E plants. Seedlings of O/E plants were infected with 105 cfu·mL−1 of AS-induced A. tumefaciens strain. Wild-type seedlings were infected with 107 cfu·mL−1 of A. tumefaciens strain as a positive control. Data are mean ± SE. * p < 0.05 compared with the wild-type by pairwise Student’s t test.
2.4. RTNLB3 and 8 Overexpression Plants were More Susceptible to P. syringae Infection
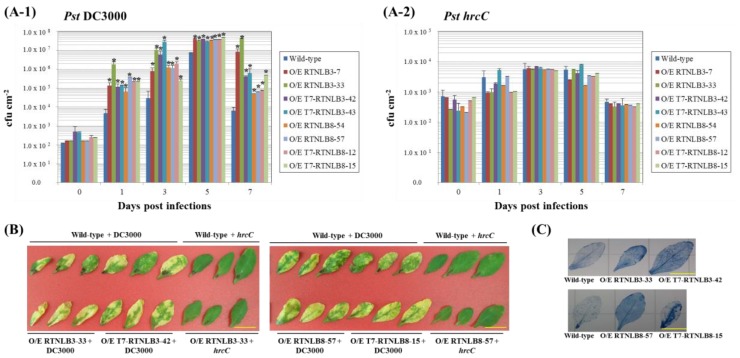
Because RTNLB1 and 2 may regulate export of FLS2, the PRR for the flagellin of Pst DC3000, to the plasma membrane during pathogen infection [40], we next examined whether overexpression of RTNLB3 or 8 could affect the plant susceptibility of P. syringae. Four- to 5-week-old plant leaves were syringe-infiltrated with wild-type Pst DC3000 and the hrcC mutant, the type III secretion system-defective bacteria, as the negative control. Bacterial growth assays were used to quantify the bacterial proliferation at 0, 1, 3, 5, and 7 days post-infection. Bacterial numbers were significantly increased in plant leaves up to 5 days after infection of wild-type Pst DC3000 and decreased at 7 days after infection (Figure 5(A-1,A-2)), which indicates successful infection with P. syringae in plants, whereas bacteria numbers only slightly increased after infection of the hrcC mutant. RTNLB3 and 8 overexpression plants had relatively higher viable bacterial numbers than wild-type plants from 1 day after infection with Pst DC3000 (Figure 5(A-1)). Overexpression of RTNLB3 or 8 in transgenic plants formed more severe disease symptoms and more chlorotic haloes than wild-type plants at 5 days after infection with Pst DC3000 (Figure 5B). As well, cell death was greater in RTNLB3 and 8 overexpression than wild-type plants (Figure 5C). On infection with the hrcC mutant, wild-type and RTNLB overexpression plants showed no difference in bacterial growth (Figure 5(A-2)) and no visible disease symptoms in leaves of both plants types (Figure 5B). These results indicate that increased expression of RTNLB3 and 8 may lead to enhanced susceptibility to Pst DC3000 infection.
Figure 5.
RTNLB3 and 8 O/E plants were more sensitive to Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) infection. Panel A, leaves of wild-type and RTNLB O/E plants were syringe-infiltrated with Pst DC3000 (Panel A-1) and hrcC mutant (Panel A-2). Bacterial numbers in infected leaves were quantified at 0, 1, 3, 5, and 7 days post-infection. Data are mean ± SE. * p < 0.05 compared with the wild-type by pairwise Student’s t test. Panel B, disease symptoms of wild-type and RTNLB O/E plant leaves 5 days after infection with Pst DC3000 or hrcC mutant. Panel C, trypan blue staining of infected leaves of wild-type and RTNLB O/E plants 5 days after infection with Pst DC3000. Yellow bar = 1 cm.
2.5. RTNLB1-4 and 8 Gene Levels Differed in Various Plant Tissues
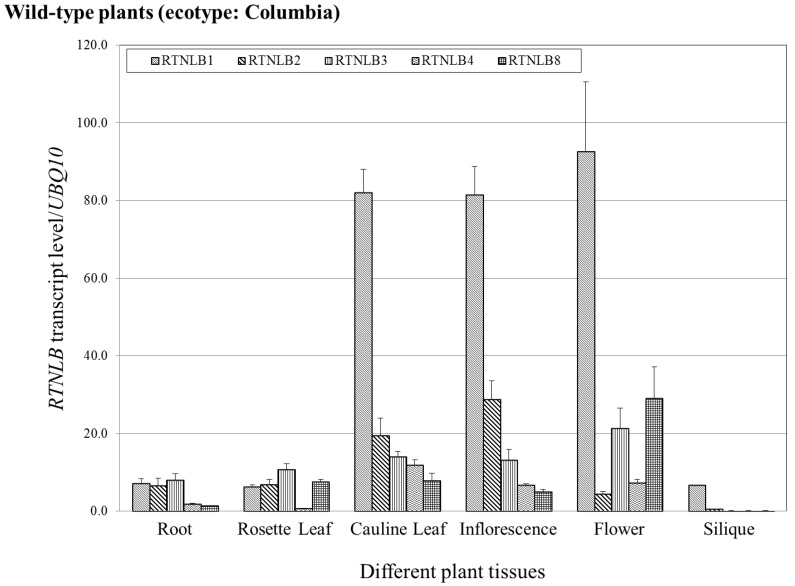
A previous study using the Arabidopsis eFP browser suggested that RTNLB1-4 and 13 genes have tissue-specific expression patterns and levels [33,44]. To understand other functions of RTNLBs in plants, we used quantitative real-time RT-PCR of RTNLB1-4 and 8 expression to determine their transcript levels in root, rosette leaf, cauline leaf, inflorescence, flower, and silique of wild-type A. thaliana (ecotype: Columbia). In roots, the transcript level was 4-fold greater for RTNLB1-3 than RTNLB4 and 8 (Figure 6). In rosette leaf, RTNLB1-3 and 8 transcripts accumulated to a similar level, whereas RTNLB4 level was the lowest among the five examined RTNLB genes (Figure 6). In cauline leaf and inflorescence, RTNLB1 transcript level was the highest, followed by RTNLB2, 3, and 4, whereas RTNLB8 level was the lowest (Figure 6). In flower, the transcript level was higher for RTNLB1 than RTNLB3 and 8, whereas RTNLB2 and 4 levels were lower than those of the other three RTNLB genes (Figure 6). In silique, RTNLB1 transcript level was the highest among the other four RTNLB genes (Figure 6). Because we generated the RTNLB3 and 8 overexpression plants in A. thaliana ecotype Wassilewskija (Ws), we also investigated RTNLB1-4 and 8 transcript levels in Ws plants. The five RTNLB transcript levels were lower in Ws plants than in Columbia plants (Figure S1). The transcript levels for RTNLB1-4 and 8 in various tissues of Ws plants differed from that in Columbia plants (Figure 6 and Figure S1). The RTNLB8 transcript level was the lowest in roots, rosette leaf, cualine leaf, inflorescence, and flower tissues of Ws plants as compared with the other four RTNLB genes (Figure S1).
Figure 6.
Levels of RTNLB1-4 and 8 in various tissues of wild-type Arabidopsis (ecotype: Columbia) plants. RNA from root, rosette leaf, cauline leaf, inflorescence, flower, and silique of wild-type plants were isolated, reverse-transcribed, and used for quantitative real-time PCR. UBQ10 (polyubiquitin 10) transcript level was an internal control. Data are mean ± SE.
3. Discussion
The reticulons (RTNLs) were first identified as endoplasmic reticulum (ER)-localized integral membrane proteins in mammalian neuron cells and have been identified in other eukaryotic cells, including yeast and plant cells [27,35]. So far, only a few members of the plant subfamily of RTNLs, named RTNLB, were demonstrated to localize in the ER and to help shape ER structures [31,32,33]. In this study, both yeast two-hybrid and GST pull-down assays revealed that RTNLB3 and 8 proteins interacted with the major component of the A. tumefaciens T-pilus, the VirB2 protein. Genetic studies have shown that the rtnlb3 and rtnlb8 single mutants were recalcitrant on A. tumefaciens-mediated transient transformation assays, whereas RTNLB3 and 8 overexpression plants were hypersusceptible to A. tumefaciens and Pseudomonas syringae pv. tomato DC3000 infection. These data suggest that RTNLB3 and 8 may play important roles in plant–microbe interactions.
Previous studies suggested that RTNLBs and the ROOT HAIR DEFECTIVE 3 (RHD3) may play a significant role in ER tubular structure formation [31,32,33,45]. RHD3 requires a functional RTNLB13 to work together on ER network alteration [46]. Additionally, in plant cells, RTNLB1-4 and 13 can interact with each other and help with ER tubular structure formation [33]. In our study, RTNLB3 and 8 protein interacted with other RTNLBs in yeast and in vitro, which further supports the possible roles of RTNLB3 and 8 in ER modeling.
Our findings show that RTNLB3 and 8 both interacted with the A. tumefaciens VirB2 protein in yeast and in vitro. Subsequently, we examined rtnlb3 and rtnlb8 mutants with a seedling-based A. tumefaciens transient transformation assay, a more sensitive method than a root-based A. tumefaciens transformation assay. Both rtnlb3 and rtnlb8 single mutants showed lower GUS activity than did wild-type seedlings. With overexpression of RTNLB3 or 8, transgenic plants were hypersensitive on both root- and seedling-based A. tumefaciens transformation assays as compared with wild-type plants, which suggests that RTNLB3 and 8 may participate in the A. tumefaciens infection process. A previous study indicated that RTNLB1, 2, and 4 may be involved in A. tumefaciens transformation [23]. RTNLB3 and/or 8 might affect A. tumefaciens infection by interacting with RTNLB1, 2 or 4, or they might have a different role during infection.
Other studies have used a co-immunoprecipitation approach to identify additional RTNLB3-interacting plant proteins and showed that RTNLB3 may also be involved in generation of ER-derived desmotubules [37,46]. Tobacco mosaic virus and Potato virus X may use desmotubules of the plant PD to spread virus particles [37,38,39] which suggests the possible roles of RTNLB3 during plant virus infection. Furthermore, RTNLB3 interacted with the vesicle-associated protein 27-1 (VAP27-1), which has high homology to the VAP33 family of SNARE-like proteins from animals, possibly involved in vesicular transport of ER [47]. RTNLB3 also interacted with a trafficking protein, synaptotagmin A (SYTA) [47]. SYTA co-localized with VAP27-1 at the ER–plasma membrane (PM) contact site to regulate endocytosis recycling at the ER–PM sites, which is related to Cabbage leaf curl virus and Tobacco mosaic virus movements [48]. A co-immunoprecipitation approach identified other RTNLB3-interacting proteins, RABA1b and RABA2c, which are members of the RAB small GTPase family and are involved in the transport between the trans-Golgi network and the PM [47,49,50]. In plant cells, RABA1b participates in the transport of de novo-synthesized FLS2, one of the PRRs for the bacterial flagellin, to the plasma membrane [51]. Most well-studied examples of pathogen-associated molecular pattern (PAMP) are the elongation factor-Tu (EF-Tu) of A. tumefaciens and the flagellin of P. syringae, which can be recognized by the elongation factor receptor (EFR) and FLS2 in plant cells, respectively [52,53,54,55]. PRRs present at the PM and can also be found at specific PM locations, in that FLS2 is enriched at PD as well [56]. A previous study demonstrated that RTNLB1 and 2 regulated ER tubular structure formation and contributed to newly synthesized FLS2 transport to the PM [40]. When the RTNLB1 protein was overexpressed or lost its function, transgenic plants and mutants were more easily infected by Pst DC3000 and showed defective FLS2-mediated immunity responses [40]. RTNLB3 or 8 overexpression plants showed increased susceptibility to Pst DC3000 infection. Therefore, RTNLB3 and 8 proteins may contribute to endomembrane trafficking in plant cells and also might participate in plant immune response by affecting plant defense response-related endomembrane trafficking pathways. Overexpression of RTNLB3 or 8 might also perturb secretion of PRRs such as FLS2 to the PM and may therefore affect transgenic plant susceptibility to phytopathogenic bacterial infection.
Although A. tumefaciens infection provokes a general defense response during early infection stages, the transfer of T-DNA and several virulence proteins into plant cells at later infection stages could significantly affect gene expression in plants, especially defense-related genes [4,57,58,59]. During A. tumefaciens initial infection, the MPK3 kinase is phosphorylated and activated, which leads to translocation of a defense-related VIP1 transcription factor into the nucleus by interacting with importin alpha and induces defense-related gene expression [21,60,61]. However, the A. tumefaciens VirE2 protein could recruit VIP1 to mediate nuclear import of T-DNA [62,63] or to sequester a low-amount of VIP1 into the cytoplasm to dampen the activity of the host defense-related response [22]. A. tumefaciens no doubt hijacks several plant proteins to overrule the plant defense response and ensure successful T-DNA transfer and expression. Although endomembrane trafficking of PRRs, including the FLS2 and EFR, is important for plant defense response perception and activation [64,65], relatively little is known about its role during A. tumefaciens infection.
In this study, we found that RTNLB3 and 8 proteins interact with VirB2 protein, the essential factor for A. tumefaciens infection and the main component of T-pilus. T-pilus is a filament structure protruding from the A. tumefaciens surface and might possibly be recognized by the host defense system. RTNLB3 and 8 might possibly participate in endomembrane trafficking of unknown receptors or the EFR protein for A. tumefaciens-induced response in plant cells. The direct link between RTNLB3/8 and endomembrane trafficking of PRRs (i.e., EFR and FLS2) awaits further investigations.
We examined RTNLB1-4 and 8 transcript levels in wild-type Arabidopsis plants (ecotype: Columbia) by real-time PCR. As compared with the expression predictions for RTNLB1-4 by using the Arabidopsis eFP browser [33,44], RTNLB1 expression was relatively higher than RTNLB2-4 and 8 in several examined tissues, which is similar to our results obtained with real-time PCR (Figure 6). However, expression levels and locations of RTNLB2-4 and 8 were slightly different from our real-time PCR results. For example, PCR results showed the highest level of RTNLB8 in flowers but Arabidopsis eFP browser results showed RTNLB8 as most abundant in cauline leaves and rosette leaves [33,44]. Our PCR results showed the level of RTNLB4 higher in cauline leaves than other tissues, but the Arabidopsis eFP browser results revealed higher level of RTNLB4 in flowers than other plant tissues [33,44]. This discrepancy might be due to two different analysis methods. The Arabidopsis eFP browser study used several microarray results from various plant cells and tissues over a long period of growth. In our results, we isolated RNA from various plant tissues of 4- to 5-week-old wild-type plants for PCR analysis. The different plant growth stages might also cause differences in results.
The different gene expression levels of endomembrane trafficking proteins affect plant growth and also have different effects on plant responses to pathogens. A member of a nuclear pore-targeting complex (PTAC), importin α (IMPα), participates in NLS cargo recognition, acts as an adaptor to bring cargo into binding of a PTAC carrier, and further interacts with nucleoporins [66,67]. Although four importin α isoforms interacted with A. tumefaciens VirD2 and VirE2 in yeast, in vitro, and in plant cells, only mutation of the importin IMPa-4 affected A. tumefaciens infection efficiency [68]. When overexpressing other importin α in the impa-4 mutant background, the mutant phenotype can be complemented, which suggests that different expression patterns and/or expression levels might affect different importin α member functions during A. tumefaciens infection [68]. Moreover, the histone H2A, HTA1 gene, is involved in T-DNA integration and T-DNA expression during A. tumefaciens infection [69,70,71]. The histone H2A family contains 13 gene members and accumulates at differing levels in roots and other plant tissues [72]. The HTA1 mutant, rat5, showed resistance to A. tumefaciens transformation efficiency. The mutant phenotype could be restored by overexpression of other HTA gene members and expression of only HTA1, not with other HTA genes, under control of its native promoter [72]. In addition, HTA1 gene expression is induced by wounding and by infection with A. tumefaciens in root cells [70,71]. These data suggest that different expression patterns of the HTA genes in various plant tissues may affect A. tumefaciens-mediated transformation efficiencies. In our results, the rtnlb8 mutant showed resistance to A. tumefaciens infection in both root- and seedling-based assays; whereas the rtnlb3 mutant showed a resistance phenotype only with the seedling-based assay, which suggests that RTNLB3 and 8 genes might play different roles in roots and seedlings during A. tumefaciens infection.
We also show RTNLB1-4 and 8 gene transcript levels in various tissues of wild-type Arabidopsis (ecotype Wassilewskija, Ws). Five RTNLB transcript levels were much lower than the same gene expression in the wild-type ecotype Columbia plants and expression patterns of the five RTNLB genes were different as well (Figure S1). Previous studies have shown that expression patterns of various members of a gene family may differ in different ecotype backgrounds [73]. For instance, the Arabidopsis sucrose transporter family contains nine members, AtSUC1-9 [73,74,75]. The AtSUC6-9 genes show high sequence homology in coding regions and in introns. From analysis of splice patterns and polymorphic sites of ATSUC6-9, AtSUC7 showed ecotype-specific splice patterns in Ws, C24, Columbia (Col-0), and Landsberg erecta (Ler) ecotypes [73]. AtSUC1 also had ecotype-specific expression and its expression was observed in the funicular epidermis of Ws, C24 and Landsberg erecta but not Col-0 [76]. So far, whether expression patterns and levels of different splice variants of RTNLB genes in plants differ is unclear. The possible roles and implications of different expression patterns of RTNLB1-4 and 8 in two Arabidopsis ecotypes (Ws and Columbia) still needs further examination.
4. Materials and Methods
4.1. Bacterial Strains and Culture
Agrobacterium tumefaciens and Escherichia coli strains used in this study are in Table S3. A. tumefaciens strains were grown in 523 medium or on 523 agar supplemented with appropriate antibiotics (rifampicin 50 μg·mL−1, gentamycin 50 μg·mL−1, and kanamycin 20 μg·mL−1; MDBio Inc., Taipei, Taiwan) at 28 °C. E. coli strains were grown at 37 °C in 2X YT medium [77] containing appropriate antibiotics (ampicillin 100 μg·mL−1, kanamycin 50 μg·mL−1).
4.2. Yeast Two-Hybrid Assays
Plasmids used for yeast two-hybrid studies are in Table S3. The bait plasmid pSST91 and the prey plasmid pGAD424 [23] were used as vectors for the construction of the various fusions. The bait plasmid pSST91 contains the LexA protein coding sequence under the control of the yeast ADH1 promoter. The prey plasmid pGAD424 generates a recombinant protein containing the GAL4 activation domain. To generate the bait plasmid expressing the LexA-RTNLB recombinant protein and the prey plasmid expressing the GAL4-RTNLB hybrid protein, EcoRI-PstI fragments of the RTNLB3, or RTNLB5-RTNLB8 coding sequences from Arabidopsis were obtained from PCR reactions by using Arabidopsis cDNA as a template, high-fidelity Phusion DNA polymerase (New England BioLabs Inc., Ipswich, MA, USA), and appropriate primers (Table S4). The PCR products were digested with EcoRI/PstI and cloned into the pSST91 or the pGAD424 as in-frame fusion to the LexA or GAL4 coding sequence.
The yeast strain CTY10-5d [23] was used for yeast two-hybrid assays. Yeast transformations involved a lithium acetate method [78]. All yeast strains were cultured at 30 °C in synthetic dropout (SD) medium [78] containing a yeast nitrogen-base, glucose, and all but the selected amino acids. The bait and prey plasmids were transformed into CTY10-5d and colonies were screened for protein interaction by colony color phenotype on SD medium with the chromogenic substrate 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) but lacking leucine and tryptophan. To quantify yeast two-hybrid interactions, the yeast liquid cultures with ortho-nitrophenyl-β-d-galactopyranosidase (ONPG) as substrates were used for β-galactosidase enzyme activity assays as described [78]. One unit of β-galactosidase enzyme activity was defined as the amount that can hydrolyze 1 μmol of ONPG to o-nitrophenol and d-galactose per minute per cell [78].
4.3. Glutathione-S-Transferase (GST) Protein Affinity Purification Assays
Plasmids and bacteria used for the GST pull-down assays are in Table S3. The plasmids pGEX4T-1 or pET42a were used to generate recombinant proteins fused in-frame with the GST tag. The plasmids pET23a and pET28a were used to express the T7-tagged fusion proteins in the E. coli strain BL21(DE3). The coding sequence of the RTNLB3 and 8 genes were amplified by PCR with Arabidopsis cDNA used as templates, high-fidelity Phusion DNA polymerase, and appropriate primers (Table S4). The PCR fragments were digested with EcoRI and PstI, which were subsequently cloned into the pBluescript plasmid. The EcoRI-NotI fragments containing RTNLB3 and 8 coding sequences were digested from the plasmids pBluescript-RTNLB3 and pBluescript-RTNLB8, respectively. The EcoRI-NotI fragments from pBluescript-RTNLB3 were cloned into pET23a or pGEX4T-1 as an in-frame fusion to the T7 tag or GST coding sequence. Similarly, the EcoRI-NotI fragments from pBluescript-RTNLB8 were cloned into pET28a or pET42a to express the T7-tagged or GST fusion proteins in bacteria. Expression and purification of GST fusion proteins and affinity purification of proteins binding to GST fusion proteins were performed as described [23,78]. The isolated protein complexes were analyzed in 12.5% SDS-polyacrylamide gels and immunoblot analysis was performed [78] with a 1:1000 dilution of anti-T7 tag primary antibody (Merck, Danvers, MA, USA) or with a 1:15,000 dilution of anti-GST primary antibody (GE Healthcare, Piscataway, NJ, USA), followed by a 1:20,000 dilution of secondary antibody horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (PerkinElmer Life and Analytical Science, Boston, MS, USA) or a 1:15,000 dilution of HRP-conjugated donkey anti-goat IgG- (Santa Cruz Biotechnology, Dallas, TX, USA), to confirm the identities of these fusion proteins. The membranes were developed by chemiluminescent detection and subjected to autoradiography.
4.4. DNA Isolation from Arabidopsis Plants and Genomic DNA PCR Analysis
The Arabidopsis T-DNA insertion mutants rtnlb3 and rtnlb5 to rtnlb10 (ecotype: Columbia CS60,000) were identified by using the SIGnAL T-DNA Express Arabidopsis Gene Mapping Tool (http://signal.salk.edu/) [79]. Seeds of rtnlb mutant plants were obtained from the Arabidopsis Biological Resource Center (ABRC; Ohio State University). Seedlings from rtnlb mutant plants were individually grown in Gamborg’s B5 medium and leaves of 3-week-old plants were used to isolate genomic DNA as described [80]. A PCR-based approach similar to that described by Alonso et al. 2003 and the SIGnAL T-DNA Express Gene Mapping Tool (http://signal.salk.edu/) was used to determine the homozygosity of Arabidopsis rtnlb mutants. Primers for genomic DNA PCR analysis are in Table S4. The PCR reaction was conducted in a 50 μL reaction volume with 2 units of GenTaq polymerase (GMbiolab Co., Taichung, Taiwan), a 2.5 mM dNTP mixture, 1× Taq polymerase reaction buffer, and 0.25 μm of the PCR primers. The PCR amplification cycle was 95 °C for 1 min (1 cycle); 94 °C for 30 s, 56 °C for 40 s, 72 °C for 1 min (30 cycles) and 72 °C for 5 min (1 cycle).
4.5. RNA Isolation from Arabidopsis Plants and RT-PCR Analysis
RNA was extracted from root and above-ground plant tissues from 4- to 5-week-old wild-type plants (ecotypes: Columbia and Wassilewskija [Ws]), the rtnlb mutant (ecotype: Columbia), and RTNLB overexpression transgenic plants (ecotype: Ws). Plant tissues were ground with a liquid nitrogen-cooled pellet pestle in a 1.5-mL Eppendorf tube. The ground materials were mixed with TRIZOL LS reagents (Total RNA Isolation Reagent for Liquid Samples from Invitrogen, Carlsbad, CA, USA) to isolate RNA according to the manufacturer’s instructions. An amount of 1–3 μg RNA was then treated with DNase I (Thermo Fisher Scientific Inc., Waltham, MA, USA) and reactions were stopped with the addition of EDTA and heat inactivation. RT-PCR involved the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc., Waltham, MA, USA) and oligo-dT primers were used to generate the first-strand cDNA products. A series of oligonucleotide primers (Table S4) was designed to amplify the sense mRNA strand of RTNLB3 and RTNLB5-RTNLB10 genes in the PCR reactions. The level of α-tubulin was an internal control in each RT-PCR reaction. The amplified products were analyzed on an agarose gel and visualized by using a UVP BioImaging System (UVP Inc., Upland, CA, USA) and quantified by using Quantity One software (Bio-Rad Laboratories Inc., Hercules, CA, USA).
An amount of 1 μg RNA samples from various tissues of wild-type plants were reverse transcribed by using the oligo-dT primer to generate cDNA. An amount of 100 ng cDNA was used for quantitative real-time PCR with the IQ2 SYBR Green Fast qPCR System Master Mix (Bio-genesis Technologies Inc., Taipei, Taiwan) in the MS3000P QPCR system (Agilent Technologies, Santa Clara, CA, USA). Another set of oligonucleotide primers (Table S4) was used to determine levels of RTNLB1-RTNLB4 and RTNLB8 genes in quantitative real-time PCR reactions. The level of UBQ10 (polyubiquitin 10) was an internal control in each quantitative real-time PCR reaction. More than 3 independent RT-PCR or real-time PCR reactions were performed with RNA samples isolated from at least 6–12 different Arabidopsis plants.
4.6. Protein Extraction from Arabidopsis Plants and Protein Gel Blot Analysis
Root tissues from 3- to 4-week-old RTNLB3 and 8 overexpression transgenic Arabidopsis plants, and wild-type plants were used to isolate proteins. Root samples were ground with liquid nitrogen and mixed with CelLytic P (Sigma Chemical Co., St. Louis, MO, USA) containing a protease inhibitor cocktail (1:100 dilution; Sigma) according to the manufacturer’s instructions. The final protein concentrations were determined by using a BCA protein assay kit (Pierce, Rockford, IL, USA) and a spectroscopy (PARADIGM Detection Platform, Beckman Coulter Inc., Indianapolis, IN, USA). Equal amounts of plant proteins were analyzed in 12.5% SDS-polyacrylamide gels and immunoblot analysis involved use of a 1:1000 dilution of T7-tag antibody (Abcam, Cambridge, UK), then a 1:20,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit IgG antibody (PerkinElmer Life and Analytical Sciences, Boston, MA, USA). The membranes were developed by chemiluminescent detection (PerkinElmer Life and Analytical Sciences, Boston, MA, USA) and subjected to autoradiography.
4.7. Generation of RTNLB3 and 8 Overexpression Arabidopsis Transgenic Plants
A binary vector, pE1798, containing a double Cauliflower mosaic virus (CaMV) 35S promoter, a nopaline synthase (Nos) terminator, and a hygromycin resistance gene (hptII) gene as a selectable marker in the T-DNA region [23] were used to overexpress RTNLB3 or 8 gene in A. thaliana transgenic plants. The KpnI-SacI fragments from the pBluescript-RTNLB3 and pBluescript-RTNLB8 were cloned into the same sites of the pE1798 plasmid (Table S3). To overexpress the T7-tagged-RTNLB3 or T7-tagged-RTNLB8 in Arabidopsis transgenic plants, the plasmids pET23a-RTNLB3 and pET28a-RTNLB8 were used as templates for PCR with the high-fidelity Phusion DNA polymerase and appropriate primers (Table S4). The PCR fragments were digested with KpnI and SacI, then cloned into the pE1798 plasmid (Table S3). These pE1978 series plasmids were separately transformed into the non-tumorigenic strain A. tumefaciens GV3101(pMP90) [81] to generate Arabidopsis overexpression plants by a floral dip method [82].
4.8. Agrobacterium Tumefaciens-Mediated Stable, Transient Root and Seedling Transformation Assays of Rtnlb Mutant Plants and Arabidopsis RTNLB Overexpression Plants
Seeds from wild-type, rtnlb mutants, and RTNLB overexpression plants were surface-sterilized and placed on Gamborg’s B5 medium (PhytoTechnology Laboratories, Carlsbad, CA) solidified with 0.75% Bactoagar (BD Biosciences, Lenexa, KS, USA) containing appropriate antibiotics (kanamycin 50 μg·mL−1 for rtnlb mutant plants and hygromycin 20 μg·mL−1 for overexpression plants). Seedlings were transferred individually to the solidified B5 medium in baby food jars without antibiotics and grown for 3–4 weeks for stable and transient root transformation assays as described [23,83].
All A. tumefaciens strains (Table S3) were cultured in 523 medium [84] with appropriate antibiotics (rifampicin 50 μg·mL−1, kanamycin 50 μg·mL−1) at 28 °C. The overnight bacterial culture was inoculated into 25 mL of 523 medium with antibiotics and grown to 109 colony forming units (cfu)·mL−1. The bacterial cells were then washed with 0.9% sodium chloride to remove antibiotics and medium. The bacterial cells were resuspended in 0.9% sodium chloride at 105, 106, or 108 cfu·mL−1 for root transformation assays.
For stable root transformation assays, root segments were cut from 3- to 4-week-old plants and transferred to solidified Murashige and Skoog medium and co-incubated with the tumorigenic strain A. tumefaciens A208 for at 22 to 24 °C for 2 days (Table S3). After co-incubation periods, root segments were separated and transferred to MS medium with antibiotic timentin (100 μg·mL−1) but lacking hormones for 1 month to score tumor formation efficiencies. For transient root transformation assays, root segments were infected with A. tumefaciens At849 containing the pBISN1 binary vector (Table S3). After 2-day co-incubation periods, root segments were placed on the callus induction medium (CIM) including timentin at 22 to 24 °C for 4 additional days. Roots were then stained with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-gluc) staining solution for 1 day at 37 °C. Roots were examined with a stereoscopic microscope to obtain transient transformation efficiencies. For root transformation assays, at least 15 different Arabidopsis plants were infected with each A. tumefaciens strain and more than 60 root segments were examined for each plant for each independent transformation assay.
The transient seedling transformation assays (Agrobacterium-mediated enhanced seedling transformation, AGROBAST) were performed as described [43] with minor modifications. The Arabidopsis seedlings were first germinated in a 6-well plate containing half-strength MS medium (pH 5.7) and 0.5% sucrose at 22 to 24 °C for 7 days. The A. tumefaciens C58C1(pTiB6S3ΔT) strain with a pBISN1 binary vector was first grown in 523 medium with the appropriate antibiotics (rifampicin 50 μg·mL−1, kanamycin 50 μg·mL−1) at 28 °C. The overnight-grown bacteria cells were further cultured for 24 h at 28 °C in acidic AB-MES medium with 200 μM acetosyringone (AS) to induce vir gene expression [84]. After AS induction, bacterial cells were washed with sterile water and resuspended in infection solution (half-strength of the MS medium [pH 5.7], one-quarter of the AB-MES medium [pH 5.5, 0.5% sucrose, and 50 μM AS]) at 107 cfu·mL−1 for seedling transformation assays. The Arabidopsis seedlings were co-incubated with AS-induced bacteria cells at 22 to 24 °C for 3 days. Seedlings were ground with liquid nitrogen and mixed with extraction buffers for fluorescent 4-methylumbelliferyl-β-d-glucuronide (MUG) assays as described [43]. The fluorescence was determined by using a 96 microplate reader (PARADIGM Detection Platform) at 365 nm excitation and 455 nm emission. The protein concentration for each protein sample was determined with a BCA protein assay kit and spectroscopy. The relative GUS activity was the fluorescence signal normalized by an equal amount of proteins. At least 10 different Arabidopsis seedlings were infected with the A. tumefaciens strain for each independent transformation assay and more than 3 independent transformation assays were performed.
4.9. Pseudomonas Syringae Infection Assays of Arabidopsis RTNLB Overexpression Plants
All P. syringae strains were grown in King’s medium B (KB medium) at 28 °C with the antibiotic rifampicin (20 μg·mL−1). After bacterial growth at 28 °C to mid to late log phase, bacterial cells were harvested, washed, and resuspended in 5 mM magnesium chloride solutions at 104 cfu·mL−1 for syringe infiltration assays. Leaves of the 4- to 5-week-old pot-grown Arabidopsis plants were infected with P. syringae strains (Table S3) by syringe infiltrations as described [85,86] with minor modifications. The abaxial side of Arabidopsis leaf was infiltrated with bacterial suspensions by using a needless syringe. To determine bacterial populations in plant leaves, leaf discs were excised from infiltrated leaves with use of a 0.6 cm2 cork borer at 0, 1, 3, 5, and 7 days after infiltration. The leaf discs were ground with use of a plastic pestle in a small amount of 5 mM magnesium chloride solutions. The bacterial suspensions were serially diluted with magnesium chloride and cultured on KB agar plates with rifampicin (20 μg·mL−1) and cycloheximide (10 μg·mL−1) to determine viable cell numbers. For Pst DC3000 infection assays, at least 15 different Arabidopsis plants were infected with bacteria for each independent infection assay and more than 3 independent infection assays were performed.
Acknowledgments
The authors thank Erh-Min Lai and Wen-Ling Deng for providing A. tumefaciens and Pseudomonas syringae strains; and the Hwang lab members for discussion and technical assistance. This research was funded by the Ministry of Science and Technology, Taiwan (MOST 105-2313-B-005-008). This research was supported in part by the Ministry of Education, Taiwan, under the ATU plan.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/2/638/s1.
Author Contributions
Fan-Chen Huang and Hau-Hsuan Hwang conceived, contributed to experiment design and wrote the manuscript. Fan-Chen Huang, Bi-Ju Fu, Yin-Tzu Liu, Yao-Ren Chang, Shin-Fei Chi, Pei-Ru Chien and Si-Chi Huang conducted experiments and analyzed data. All authors read and approved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gelvin S.B. Plant proteins involved in Agrobacterium-mediated genetic transformation. Annu. Rev. Phytopathol. 2010;48:45–68. doi: 10.1146/annurev-phyto-080508-081852. [DOI] [PubMed] [Google Scholar]
- 2.Gelvin S.B. Traversing the cell: Agrobacterium T-DNA’s journey to the host genome. Front. Plant Sci. 2012;3:52. doi: 10.3389/fpls.2012.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lacroix B., Citovsky V. The roles of bacterial and host plant factors in Agrobacterium-mediated genetic transformation. Int. J. Biochem. Cell Biol. 2013;57:467–481. doi: 10.1387/ijdb.130199bl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pitzschke A. Infection and plant defense-transformation success hangs by a thread. Front. Plant Sci. 2013;4:519. doi: 10.3389/fpls.2013.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gohlke J., Deeken R. Plant responses to Agrobacterium tumefaciens and crown gall development. Front. Plant Sci. 2014;5:155. doi: 10.3389/fpls.2014.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brencic A., Winans S.C. Detection of and response to signals involved in host–microbe interactions by plant-associated bacteria. Microbiol. Mol. Biol. Rev. 2005;69:155–194. doi: 10.1128/MMBR.69.1.155-194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCullen C.A., Binns A.N. Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu. Rev. Cell Dev. Biol. 2006;22:101–127. doi: 10.1146/annurev.cellbio.22.011105.102022. [DOI] [PubMed] [Google Scholar]
- 8.Zechner E.L., Lang S., Schildbach J.F. Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R. Soc. B Biol. Sci. 2012;367:1073–1087. doi: 10.1098/rstb.2011.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatty M., Laverde Gomez J.A., Christie P.J. The expanding bacterial type IV secretion lexicon. Res. Microbiol. 2013;164:620–639. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandran V. Type IV secretion machinery: Molecular architecture and function. Biochem. Soc. Trans. 2013;41:17–28. doi: 10.1042/BST20120332. [DOI] [PubMed] [Google Scholar]
- 11.Christie P.J., Whitaker N., Gonzalez-Rivera C. Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta. 2014;1843:1578–1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallden K., Rivera-Calzada A., Waksman G. Type IV secretion systems: Versatility and diversity in function. Cell Microbiol. 2010;12:1203–1212. doi: 10.1111/j.1462-5822.2010.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waksman G., Fronzes R. Molecular architecture of bacterial type IV secretion systems. Trends Biochem. Sci. 2010;35:691–698. doi: 10.1016/j.tibs.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Waksman G., Orlova E.V. Structural organisation of the type IV secretion systems. Curr. Opin. Microbiol. 2014;17:24–31. doi: 10.1016/j.mib.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai E.M., Kado C.I. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 1998;180:2711–2717. doi: 10.1128/jb.180.10.2711-2717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai E.M., Chesnokova O., Banta L.M., Kado C.I. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J. Bacteriol. 2000;182:3705–3716. doi: 10.1128/JB.182.13.3705-3716.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai E.M., Eisenbrandt R., Kalkum M., Lanka E., Kado C.I. Biogenesis of T pili in Agrobacterium tumefaciens requires precise VirB2 propilin cleavage and cyclization. J. Bacteriol. 2002;184:327–330. doi: 10.1128/JB.184.1.327-330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenbrandt R., Kalkum M., Lai E.M., Lurz R., Kado C.I., Lanka E. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt-Eisenlohr H., Domke N., Angerer C., Wanner G., Zambryski P.C., Baron C. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 1999;181:7485–7492. doi: 10.1128/jb.181.24.7485-7492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aly K.A., Baron C. The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens. Microbiology. 2007;153:3766–3775. doi: 10.1099/mic.0.2007/010462-0. [DOI] [PubMed] [Google Scholar]
- 21.Djamei A., Pitzschke A., Nakagami H., Rajh I., Hirt H. Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science. 2007;318:453–456. doi: 10.1126/science.1148110. [DOI] [PubMed] [Google Scholar]
- 22.Shi Y., Lee L.Y., Gelvin S.B. Is VIP1 important for Agrobacterium-mediated transformation? Plant J. 2014;79:848–860. doi: 10.1111/tpj.12596. [DOI] [PubMed] [Google Scholar]
- 23.Hwang H.H., Gelvin S.B. Plant proteins that interact with VirB2, the Agrobacterium pilin protein, mediate plant transformation. Plant Cell. 2004;16:3148–3167. doi: 10.1105/tpc.104.026476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roebroek A.J., van de Velde H.J., Van Bokhoven A., Broers J.L., Ramaekers F.C., Van de Ven W.J. Cloning and expression of alternative transcripts of a novel neuroendocrine-specific gene and identification of its 135-kDa translational product. J. Biol. Chem. 1993;268:13439–13447. [PubMed] [Google Scholar]
- 25.Van de Velde H.J., Senden N.H., Roskams T.A., Broers J.L., Ramaekers F.C., Roebroek A.J., Van de Ven W.J. NSP-encoded reticulons are neuroendocrine markers of a novel category in human lung cancer diagnosis. Cancer Res. 1994;54:4769–4776. [PubMed] [Google Scholar]
- 26.Senden N.H., Timmer E.D., Boers J.E., van de Velde H.J., Roebroek A.J., Van de Ven W.J., Broers J.L., Ramaekers F.C. Neuroendocrine-specific protein C (NSP-C): Subcellular localization and differential expression in relation to NSP-A. Eur. J. Cell Biol. 1996;69:197–213. [PubMed] [Google Scholar]
- 27.Oertle T., Schwab M.E. Nogo and its paRTNers. Trends Cell Biol. 2003;13:187–194. doi: 10.1016/S0962-8924(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 28.Voeltz G.K., Prinz W.A., Shibata Y., Rist J.M., Rapoport T.A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 29.Hu J., Shibata Y., Voss C., Shemesh T., Li Z., Coughlin M., Kozlov M.M., Rapoport T.A., Prinz W.A. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- 30.Nziengui H., Schoefs B. Functions of reticulons in plants: What we can learn from animals and yeasts. Cell Mol. Life Sci. 2009;66:584–595. doi: 10.1007/s00018-008-8373-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tolley N., Sparkes I.A., Aunter P.R., Craddock C.P., Nuttall J., Roberts L.M., Hawes C., Pedrazzini E., Frigerio L. Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic. 2008;9:94–102. doi: 10.1111/j.1600-0854.2007.00670.x. [DOI] [PubMed] [Google Scholar]
- 32.Tolley N., Sparkes I., Craddock C.P., Eastmond P.J., Runions J., Hawes C., Frigerio L. Transmembrane domain length is responsible for the ability of a plant reticulon to shape endoplasmic reticulum tubules in vivo. Plant J. 2010;64:411–418. doi: 10.1111/j.1365-313X.2010.04337.x. [DOI] [PubMed] [Google Scholar]
- 33.Sparkes I., Tolley N., Aller I., Svozil J., Osterrieder A., Botchway S., Mueller C., Frigerio L., Hawes C. Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell. 2010;22:1333–1343. doi: 10.1105/tpc.110.074385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oertle T., Klinger M., Stuermer C.A., Schwab M.E. A reticular rhapsody: Phylogenic evolution and nomenclature of the RTN/Nogo gene family. FASEB J. 2003;17:1238–1247. doi: 10.1096/fj.02-1166hyp. [DOI] [PubMed] [Google Scholar]
- 35.Nziengui H., Bouhidel K., Pillon D., Der C., Marty F., Schoefs B. Reticulon-like proteins in Arabidopsis thaliana: Structure organization and ER localization. FEBS Lett. 2007;581:3356–3362. doi: 10.1016/j.febslet.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 36.Marmagne A., Rouet M.A., Ferro M., Rolland N., Alcon C., Joyard J., Garin J., Barbier-Brygoo H., Ephritikhine G. Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol. Cell Proteomics. 2004;3:675–691. doi: 10.1074/mcp.M400001-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Knox K., Wang P., Kriechbaumer V., Tilsner J., Frigerio L., Sparkes I., Hawes C., Oparka K.J. Putting the squeeze on plasmodesmata: A role for reticulons in primary plasmodesmata formation. Plant Physiol. 2015;168:1563–1572. doi: 10.1104/pp.15.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu C.H., Lee S.C., Wang C.W. Viral protein targeting to the cortical endoplasmic reticulum is required for cell-cell spreading in plants. J. Cell Biol. 2011;193:521–535. doi: 10.1083/jcb.201006023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tilsner J., Linnik O., Louveaux M., Roberts I.M., Chapman S.N., Oparka K.J. Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 2013;201:981–995. doi: 10.1083/jcb.201304003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee H.Y., Bowen C.H., Popescu G.V., Kang H.G., Kato N., Ma S., Dinesh-Kumar S., Snyder M., Popescu S.C. Arabidopsis RTNLB1 and RTNLB2 Reticulon-like proteins regulate intracellular trafficking and activity of the FLS2 immune receptor. Plant Cell. 2011;23:3374–3391. doi: 10.1105/tpc.111.089656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mysore K.S., Bassuner B., Deng X.B., Darbinian N.S., Motchoulski A., Ream W., Gelvin S.B. Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol. Plant Microbe Interact. 1998;11:668–683. doi: 10.1094/MPMI.1998.11.7.668. [DOI] [PubMed] [Google Scholar]
- 42.Mysore K.S., Kumar C.T., Gelvin S.B. Arabidopsis ecotypes and mutants that are recalcitrant to Agrobacterium root transformation are susceptible to germ-line transformation. Plant J. 2000;21:9–16. doi: 10.1046/j.1365-313x.2000.00646.x. [DOI] [PubMed] [Google Scholar]
- 43.Wu H.Y., Liu K.H., Wang Y.C., Wu J.F., Chiu W.L., Chen C.Y., Wu S.H., Sheen J., Lai E.M. AGROBEST: An efficient Agrobacterium-mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods. 2014;10:19. doi: 10.1186/1746-4811-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. An ’Electronic fluorescent pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J., Stefano G., Brandizzi F., Zheng H. Arabidopsis RHD3 mediates the generation of the tubular ER network and is required for Golgi distribution and motility in plant cells. J. Cell Sci. 2011;124:2241–2252. doi: 10.1242/jcs.084624. [DOI] [PubMed] [Google Scholar]
- 46.Lee H., Sparkes I., Gattolin S., Dzimitrowicz N., Roberts L.M., Hawes C., Frigerio L. An Arabidopsis reticulon and the atlastin homologue RHD3-like2 act together in shaping the tubular endoplasmic reticulum. New Phytol. 2013;197:481–489. doi: 10.1111/nph.12038. [DOI] [PubMed] [Google Scholar]
- 47.Kriechbaumer V., Botchway S.W., Slade S.E., Knox K., Frigerio L., Oparka K., Hawes C. Reticulomics: Protein-protein interaction studies with two plasmodesmata-localized reticulon family proteins identify binding partners enriched at plasmodesmata, endoplasmic reticulum, and the plasma membrane. Plant Physiol. 2015;169:1933–1945. doi: 10.1104/pp.15.01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis J.D., Lazarowitz S.G. Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc. Natl. Acad. Sci. USA. 2010;107:2491–2496. doi: 10.1073/pnas.0909080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feraru E., Feraru M.I., Asaoka R., Paciorek T., De Rycke R., Tanaka H., Nakano A., Friml J. BEX5/RabA1b regulates trans-Golgi network-to-plasma membrane protein trafficking in Arabidopsis. Plant Cell. 2012;24:3074–3086. doi: 10.1105/tpc.112.098152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asaoka R., Uemura T., Ito J., Fujimoto M., Ito E., Ueda T., Nakano A. Arabidopsis RABA1 GTPases are involved in transport between the trans-Golgi network and the plasma membrane, and are required for salinity stress tolerance. Plant J. 2013;73:240–249. doi: 10.1111/tpj.12023. [DOI] [PubMed] [Google Scholar]
- 51.Choi S.W., Tamaki T., Ebine K., Uemura T., Ueda T., Nakano A. RABA members act in distinct steps of subcellular trafficking of the FLAGELLIN SENSING2 receptor. Plant Cell. 2013;25:1174–1187. doi: 10.1105/tpc.112.108803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 2006;18:465–476. doi: 10.1105/tpc.105.036574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 54.Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 55.Couto D., Zipfel C. Regulation of pattern recognition receptor signaling in plants. Nat. Rev. Immunol. 2016;16:537–552. doi: 10.1038/nri.2016.77. [DOI] [PubMed] [Google Scholar]
- 56.Beck M., Heard W., Mbengue M., Robatzek S. The INs and OUTs of pattern recognition receptors at the cell surface. Plant Biol. 2012;15:367–374. doi: 10.1016/j.pbi.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 57.Ditt R.F., Nester E., Comai L. The plant cell defense and Agrobacterium tumefaciens. FEMS Microbiol. Lett. 2005;247:207–213. doi: 10.1016/j.femsle.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 58.Ditt R.F., Nester E.W., Comai L. Plant gene expression response to Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA. 2001;98:10954–10959. doi: 10.1073/pnas.191383498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Veena, Doerge R.W., Gelvin S.B. Transfer of T-DNA and Vir proteins to plant cells by Agrobacterium tumefaciens induces expression of host genes involved in mediating transformation and suppresses host defense gene expression. Plant J. 2003;35:219–236. doi: 10.1046/j.1365-313x.2003.01796.x. [DOI] [PubMed] [Google Scholar]
- 60.Pitzschke A., Djamei A., Teige M., Hirt H. VIP1 response elements mediate mitogen-activated protein kinase 3-induced stress gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:18414–18419. doi: 10.1073/pnas.0905599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pitzschke A., Hirt H. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J. 2010;29:1021–1032. doi: 10.1038/emboj.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Citovsky V., Kapelnikov A., Oliel S., Zakai N., Rojas M.R., Gilbertson R.L., Tzfira T., Loyter A. Protein interactions involved in nuclear import of the Agrobacterium VirE2 protein in vivo and in vitro. J. Biol. Chem. 2004;279:29528–29533. doi: 10.1074/jbc.M403159200. [DOI] [PubMed] [Google Scholar]
- 63.Tzfira T., Vaidya M., Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001;20:3596–3607. doi: 10.1093/emboj/20.13.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ben Khaled S., Postma J., Robatzek S. A moving view: Subcellular trafficking processes in pattern recognition receptor-triggered plant immunity. Annu. Rev. Phytopathol. 2015;53:379–402. doi: 10.1146/annurev-phyto-080614-120347. [DOI] [PubMed] [Google Scholar]
- 65.Gu Y., Zavaliev R., Dong X. Membrane trafficking in plant immunity. Mol. Plant. 2017;10:1026–1034. doi: 10.1016/j.molp.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gorlich D., Kostka S., Kraft R., Dingwall C., Laskey R.A., Hartmann E., Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 1995;5:383–392. doi: 10.1016/S0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 67.Bednenko J., Cingolani G., Gerace L. Importin beta contains a COOH-terminal nucleoporin binding region important for nuclear transport. J. Cell Biol. 2003;162:391–401. doi: 10.1083/jcb.200303085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bhattacharjee S., Lee L.Y., Oltmanns H., Cao H., Cuperus J., Gelvin S.B. AtImpa-4, an Arabidopsis importin α isoform, is preferentially involved in Agrobacterium-mediated plant transformation. Plant Cell. 2008;20:2661–2680. doi: 10.1105/tpc.108.060467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mysore K.S., Nam J., Gelvin S.B. An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. Proc. Natl. Acad. Sci. USA. 2000;97:948–953. doi: 10.1073/pnas.97.2.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yi H., Mysore K.S., Gelvin S.B. Expression of the Arabidopsis histone H2A-1 gene correlates with susceptibility to Agrobacterium transformation. Plant J. 2002;32:285–298. doi: 10.1046/j.1365-313X.2002.01425.x. [DOI] [PubMed] [Google Scholar]
- 71.Tenea G.N., Spantzel J., Lee L.Y., Zhu Y., Lin K., Johnson S.J., Gelvin S.B. Overexpression of several Arabidopsis histone genes increases Agrobacterium-mediated transformation and transgene expression in plants. Plant Cell. 2009;21:3350–3367. doi: 10.1105/tpc.109.070607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yi H., Sardesai N., Fujinuma T., Chan C.W., Gelvin S.B. Constitutive expression exposes functional redundancy between the Arabidopsis histone H2A gene HTA1 and other H2A gene family members. Plant Cell. 2006;18:1575–1589. doi: 10.1105/tpc.105.039719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sauer N., Ludwig A., Knoblauch A., Rothe P., Gahrtz M., Klebl F. AtSUC8 and AtSUC9 encode functional sucrose transporters, but the closely related AtSUC6 and AtSUC7 genes encode aberrant proteins in different Arabidopsis ecotypes. Plant J. 2004;40:120–130. doi: 10.1111/j.1365-313X.2004.02196.x. [DOI] [PubMed] [Google Scholar]
- 74.Lemoine R. Sucrose transporters in plants: Update on function and structure. Biochimica et Biophysica Acta. 2000;1465:246–262. doi: 10.1016/S0005-2736(00)00142-5. [DOI] [PubMed] [Google Scholar]
- 75.Williams L.E., Lemoine R., Sauer N. Sugar transporters in higher plants—A diversity of roles and complex regulation. Trends Plant Sci. 2000;5:283–290. doi: 10.1016/S1360-1385(00)01681-2. [DOI] [PubMed] [Google Scholar]
- 76.Feuerstein A., Niedermeier M., Bauer K., Engelmann S., Hoth S., Stadler R., Sauer N. Expression of the AtSUC1 gene in the female gametophyte, and ecotype-specific expression differences in male reproductive organs. Plant Biol. 2010;12:105–114. doi: 10.1111/j.1438-8677.2010.00389.x. [DOI] [PubMed] [Google Scholar]
- 77.Sambrook J., Russell D.W. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2001. [Google Scholar]
- 78.Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. Current Protocols in Molecular Biology. John Wiley & Sons Inc.; Hoboken, NJ, USA: 2003. [Google Scholar]
- 79.Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 80.Dellaporta S.L., Wood J., Hicks J.B. A plant DNA minipreperation: Version 2. Plant Mol. Biol. Rep. 1983;1:19–22. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- 81.Koncz C., Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 1986;204:383–396. doi: 10.1007/BF00331014. [DOI] [Google Scholar]
- 82.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 83.Zhu Y., Nam J., Humara J.M., Mysore K.S., Lee L.Y., Cao H., Valentine L., Li J., Kaiser A.D., Kopecky A.L., et al. Identification of Arabidopsis rat mutants. Plant Physiol. 2003;132:494–505. doi: 10.1104/pp.103.020420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hwang H.H., Wang M.H., Lee Y.L., Tsai Y.L., Li Y.H., Yang F.J., Liao Y.C., Lin S.K., Lai E.M. Agrobacterium-produced and exogenous cytokinin-modulated Agrobacterium-mediated plant transformation. Mol. Plant Pathol. 2010;11:677–690. doi: 10.1111/j.1364-3703.2010.00637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Deng W.L., Preston G., Collmer A., Chang C.J., Huang H.C. Characterization of the hrpC and hrpRS operons of Pseudomonas syringae pathovars syringae, tomato, and glycinea and analysis of the ability of hrpF, hrpG, hrcC, hrpT, and hrpV mutants to elicit the hypersensitive response and disease in plants. J. Bacteriol. 1998;180:4523–4531. doi: 10.1128/jb.180.17.4523-4531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Katagiri F., Thilmony R., He S.Y. The Arabidopsis thaliana-Pseudomonas syringae interaction. Arabidopsis Book. 2002;1:e0039. doi: 10.1199/tab.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.