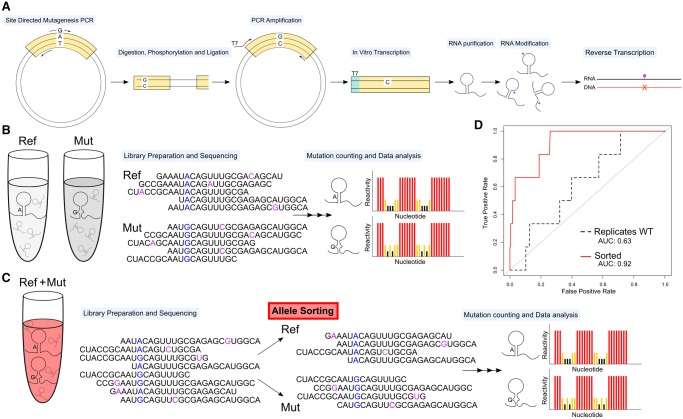
FIGURE 2.
High-throughput strategy to identify riboSNitches is improved by clone-free allele-specific sorting. (A) We performed site-directed mutagenesis followed by SHAPE-MaP. (B) In a traditional experiment, reference and mutant RNAs are modified and prepared for sequencing in separate tubes. (C) In our novel protocol, reference and mutant simultaneously undergo modification. The sequencing reads are sorted based on the reference/mutant allele and SHAPE reactivities are calculated from the sorted reads. (D) By simulating the traditional experimental approach, we have poor differentiation (AUC 0.63, dotted line), while simulating our novel within sample approach results in a dramatic improvement (AUC 0.92, red line).