FIGURE 7.
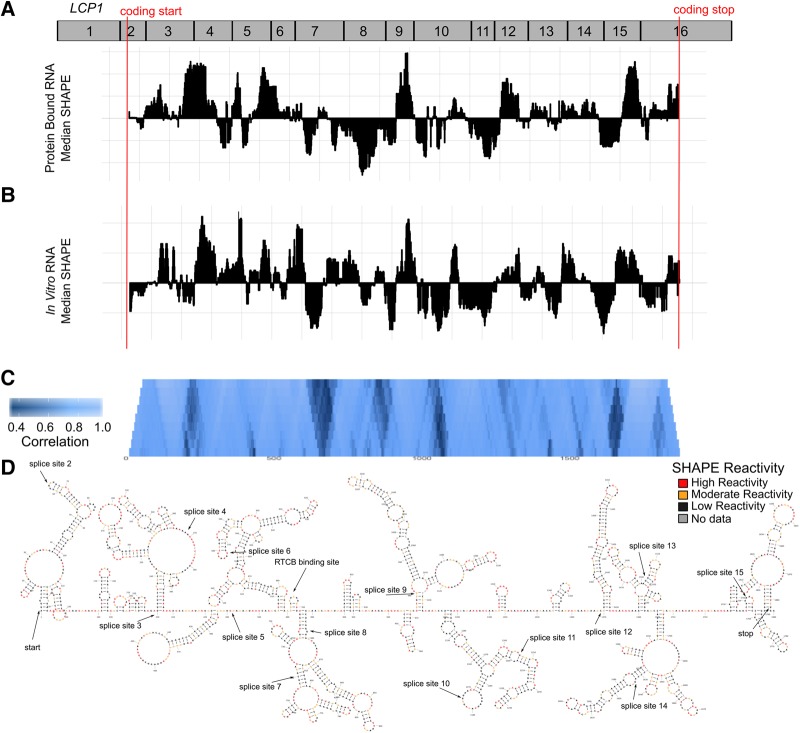
LCP1 mRNA secondary structures are similar in the presence and absence of protein. (A) LCP1 RNPs were probed and analyzed to generate median reactivity data. Low SHAPE regions indicate likely structured regions, while high SHAPE regions are likely unstructured. The median reactivity within the CDS of LCP1 corresponds closely with the pattern of (B) naked RNA reactivity data. (C) The pattern of SHAPE reactivity is highly correlated (>0.82 overall). Correlations are performed over multiple windows, from 10 to 100, and lighter blue indicates higher correlation. Various regions of difference can be seen throughout the CDS as darker bands. (D) LCP1 secondary structure model for the entire 1884 nt CDS using SHAPE data from unbound RNA to direct RNA structure prediction. Regions of interest are labeled, including splice junctions and potential RNA-binding protein sites.