Significance
We demonstrate the genetic modification of structural color in a living system by using bacteria Iridescent 1 (IR1) as a model system. IR1 colonies consist of rod-shaped bacteria that pack in a dense hexagonal arrangement through gliding and growth, thus interfering with light to give a bright, green, and glittering appearance. By generating IR1 mutants and mapping their optical properties, we show that genetic alterations can change colony organization and thus their visual appearance. The findings provide insight into the genes controlling structural color, which is important for evolutionary studies and for understanding biological formation at the nanoscale. At the same time, it is an important step toward directed engineering of photonic systems from living organisms.
Keywords: genetics, structural color, Flavobacteria, self-organization, disorder
Abstract
Naturally occurring photonic structures are responsible for the bright and vivid coloration in a large variety of living organisms. Despite efforts to understand their biological functions, development, and complex optical response, little is known of the underlying genes involved in the development of these nanostructures in any domain of life. Here, we used Flavobacterium colonies as a model system to demonstrate that genes responsible for gliding motility, cell shape, the stringent response, and tRNA modification contribute to the optical appearance of the colony. By structural and optical analysis, we obtained a detailed correlation of how genetic modifications alter structural color in bacterial colonies. Understanding of genotype and phenotype relations in this system opens the way to genetic engineering of on-demand living optical materials, for use as paints and living sensors.
Besides pigmentation, nature’s palette comprises colors that can be achieved by nanostructuring materials at the scale of visible light wavelengths. In this way, living organisms are able to modify their optical appearance (in terms of color and angular dependency) with a large degree of freedom (1–3). As an example, while vivid, iridescent colors are obtained from light interacting with periodically arranged scattering elements, less angle-dependent colors rely on quasi-ordered (4) or completely random structures (5, 6). Such structural colors are found in a large variety of organisms spanning all kingdoms of life, from eukaryotes (1, 3, 7) to prokaryotes (8, 9). In many species, the biological significance of structural coloration represents an evolutionary advantage for camouflage (10), sexual selection (11), thermal regulation (12), photosynthesis (13, 14), and intraspecies signaling (15, 16). Despite such a variety of mechanisms and species displaying structural coloration, there is still little knowledge on the processes regulating the development of such colors in any living system (17–20) in terms of genotype–phenotype relation, which is key to the understanding of development, function, and evolution of structural colors (21).
In this work, Flavobacterium strain Iridescent 1 (IR1) was used as a model system. Flavobacteria are widely distributed, Gram-negative, biopolymer-degrading bacteria. Their motility is via gliding, where cells move over a surface in a pili-independent, flagella-independent manner using the proton motive force to generate traction via a novel molecular motor (22). IR1 colonies display vivid, bright structural coloration, similar to the gliding bacteria from other Cytophaga–Flavobacterium–Bacteroides phyla (8, 9, 23). Through transposon insertions in the WT strain, thus creating a library of genetic variants with nonessential genes knocked out, we were able to select mutants displaying different optical properties and subsequently map the genes responsible against the sequenced IR1 genome. The structural and optical characterizations of WT and mutant colonies were combined with a genetic study of the pathways responsible for the spatial organization of the bacteria. This coordinated study of genotype–phenotype relation provides an unparalleled insight into the genetic pathways responsible for structural colors from living organisms. Furthermore, by analyzing the development of structural colors in different growing conditions and substrates (including algae and other biotic surfaces) we suggest where bacterial colonies may exhibit structural color in their natural environment.
Overview of Flavobacterium Strain and Mutants
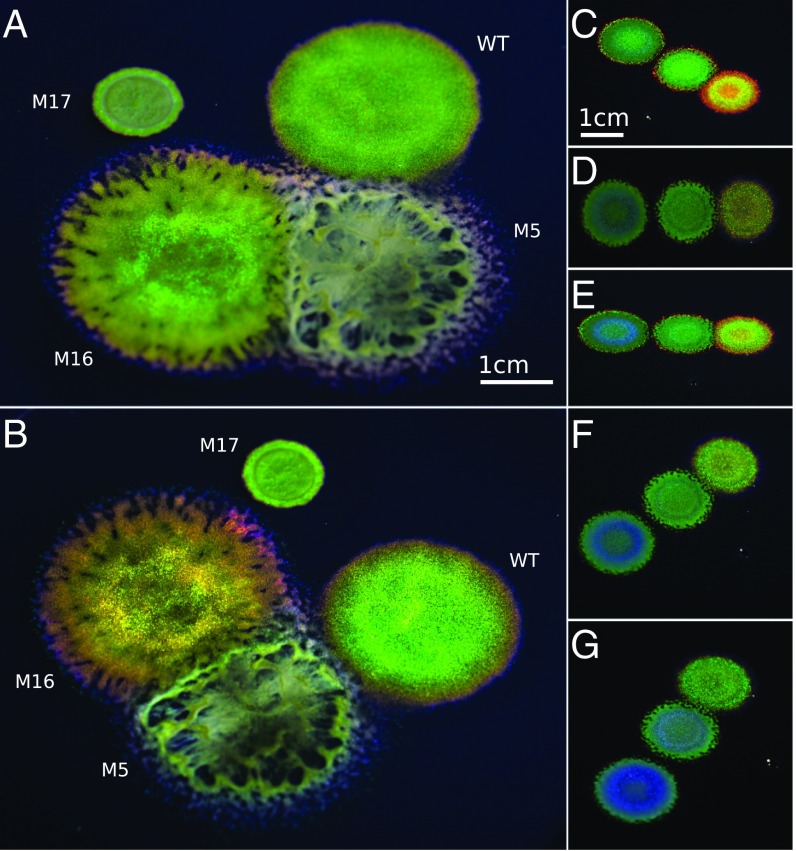
The bacterial strain IR1 was isolated during a screening of estuarine sediment samples from the Neckarhaven region of Rotterdam Harbor, The Netherlands. The cultured strains of Flavobacteria most closely related to IR1 were Flavobacterium aquidurense and Flavobacterium pectinovorum with 99% identity on the basis of 16S sequence comparison (Materials and Methods and SI Appendix, Fig. S1). F. aquidurense was originally isolated from a freshwater creek in Germany (24) and F. pectinovorum from soil in the United Kingdom (25). Neither bacterium has been reported to display structural coloration. On Artificial Sea Water Black Carrageenan (ASWBC) agar plates (see SI Appendix, Tables S1 and S2 for definitions), the WT IR1 strain showed gliding motility and displayed a bright, brilliant green structural coloration as seen in Fig. 1 A and B. Libraries of mutants were generated using the HiMar transposon and screened for altered optical properties. Strains with different optical properties (ranging from intense red to blue colors to mutations with reduced or no coloration) were isolated (Fig. 1 A–G). The WT strain and three mutants labeled M5, M16, and M17 were selected as representative models to study the interplay between motility, cell geometry, and other aspects underpinning structural organization of the bacterial colonies (Fig. 1 A and B). M5 had comparable motility and cell shape to the WT but failed to provide clear structural coloration under the same growing conditions. Colonies of M16 had a very intense, red-shifted appearance. M17 displayed decreased motility compared with WT and a barely visible angle dependency. An overview of the physical parameters of key mutants and the mapping of transposon insertions leading to these phenotypes is provided in SI Appendix, Table S3.
Fig. 1.
Structural coloration of Flavobacterium IR1 WT and mutants. (A and B) Photographs of WT and three mutants, grown for 4 d, taken at two different angles to exhibit the colonies’ pronounced angle-dependent coloration. WT and M16 produce vivid coloration whereas M17 produces some coloration and M5 very little in comparison. (Scale bars in A and B, 1 cm.) (C–G) Mutants M22, M65, and M41 (left to right in all photographs) photographed from five different angles under the same illumination showing the variation in color after growth for 2 d. D is photographed from directly above, whereas the other observation angles are oblique. (Scale bars in C–G, 1 cm.)
Differences in Organizational Capacity Are Revealed by Electron Microscopy
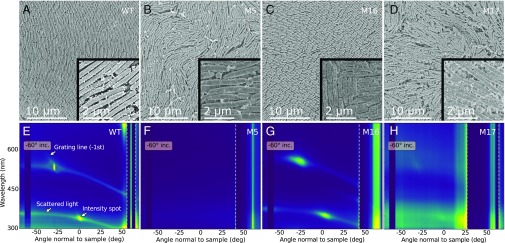
To gain insight into the structural arrangement causing the coloration, we performed structural characterization by scanning electron microscopy (SEM) on fixed colonies (Fig. 2 A–D). Studies by SEM were facilitated by a fixation procedure that resulted in the dried material maintaining a structural color (SI Appendix, Fig. S10). A clear difference in bacterial cell geometry between strains was found and the trend was confirmed by statistically significant measurements of cell lengths and diameters based on several images (SI Appendix, Fig. S6 A–H). The WT colony had a significantly lower variation in cell diameter than the mutant colonies, which possibly facilitates the packing into a regular lattice. The length of the WT bacteria is comparable in size and variation to M5, while M16 and M17 were shorter and longer than the WT, respectively. M17, despite being significantly longer than the WT, stacked in defined layers like the WT and M16—albeit less densely. M5, however, was not as well organized as the WT and the bacteria were poorly aligned with respect to their neighboring cells (Fig. 2B).
Fig. 2.
Structural and optical investigations of IR1 strains. (A–D) SEM top view images of the four bacterium strains WT, M5, M16, and M17, at identical magnifications. (E–H) Scattered intensity for samples upon light illumination with an incident angle of . To visualize the specular reflections, the signals within the dashed lines are divided by 900, 200, 350, and 250, respectively, from Left to Right. In E, text annotations highlight the features caused by structural organization: confined intensity spots lying on a very weak line governed by the grating equation (stated in SI Appendix), as well as a more defined scattered line spanning the whole angular range and also crossing the diffraction spots.
The regularity of the cells within these colonies was investigated by calculating the spatial autocorrelation of the SEM images (SI Appendix, Fig. S7). The profiles of the autocorrelation, shown in SI Appendix, Fig. S7 E–H, provided good estimations of the packing order of the bacteria. A particularly long-ranged correlation was observed in WT and M16, indicating that the relative position and orientation of bacteria were maintained over tens of micrometers. M5 and M17 showed little or no correlation. This difference in packing order was predicted to strongly influence the optical response of the bacterial colonies, as discussed in the following section. From the autocorrelation analysis, the periodicities of the bacterial arrangements were estimated to be , , and for WT, M5, and M16, respectively (Materials and Methods and SI Appendix, Fig. S7 E–G). For M17 it was not possible to extract a meaningful estimate due to lack of a clear peak in the autocorrelation spectrum, indicating a lack of ability to organize in a regular pattern (SI Appendix, Fig. S7L).
Photonic Responses of Colonies Are Closely Related to Local Colony Organization
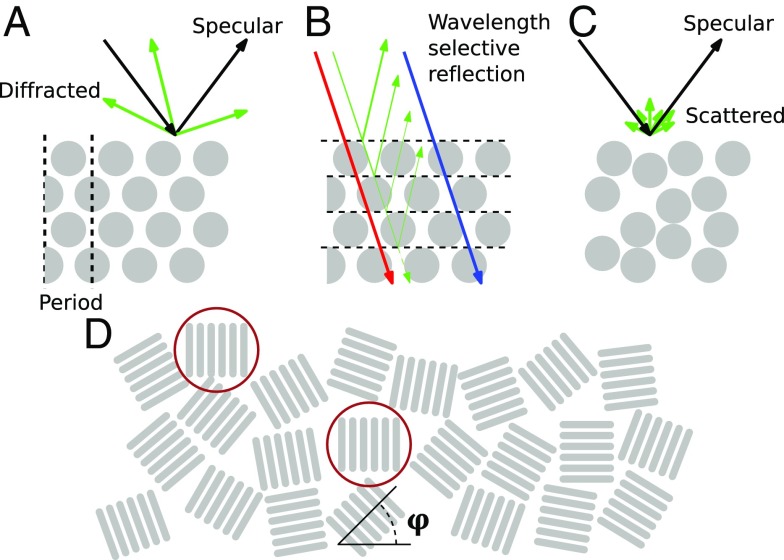
The WT and selected mutants cultured on ASWBC agar were studied with an optical goniometer. Fig. 2 E–H shows the scattering behavior of the different bacterial strains when illuminated at a grazing incidence angle of with respect to the normal of the surface, with the complete set of measurements for different incident angles reported in SI Appendix, Fig. S3. For all colonies, a strong specular reflection arising from the air interface is visible at , which is mainly dominated by the reflection from the agar. We therefore focused our analysis on the scattering properties of the colonies at nonspecular angles. In fact, the periodic arrangement of the bacteria dramatically affects the visual appearance of the colony. In the case of perfect packing (where the bodies of the bacteria form a 2D close-packed hexagonal lattice as schematically shown in Fig. 3A), the colony diffracts light only at angles specified by the grating equation (SI Appendix). The intensity profile of the diffracted light depends on the lattice constant and the packing geometry, resulting in brighter color-selective reflections at specific angles (Fig. 3 A and B). For example, the grating equation and the scale invariance of Maxwell’s equations imply that a larger lattice constant compresses the angular distribution of diffraction spots and gives longer peak reflection wavelengths.
Fig. 3.
Diagrams explaining the light scattering observed from colonies of IR1. (A) If the cells are arranged in a strictly periodic manner, they will reflect light only at certain angles, but with a very strong intensity as described by the grating equation (Materials and Methods). (B) The intense color selectivity is determined by the layered stacking of the bacteria, where interference, for example, reflects green light but transmits blue and red. (C) If this periodicity is disrupted, the grating effect is obscured, and less strong intensity of reflected light is observed, but over a broader angular range. (D) Top view of bacteria. Since the bacteria macroscopically do not have a preferred orientation with respect to in-plane rotation (), the visual appearance is independent of rotation along this axis. For example, only the two encircled, aligned areas will contribute to the optical response in the observation plane perpendicular to their orientation.
Deviations from the ideal, periodic structure can be grouped in two categories: (i) disorder in the packing geometry, depicted in Fig. 3C, and (ii) disorder in domains with the same orientation of bacteria, shown in Fig. 3D. The first effect is the dominant contribution to the scattered light around the two diffraction spots at and in Fig. 2E. The latter effect makes the scattering signature independent of in-plane rotation (SI Appendix, Fig. S4).
The diffraction spots for the WT strain are clearly distinguishable (Fig. 2E) and in agreement with the grating equation (SI Appendix, Fig. S3). Their broadening in both angle and wavelength can be attributed to the variation of the bacteria diameter. Such packing gives rise to a glittering, angle-dependent appearance of the bacteria strains, as was previously observed by Kientz et al. (26, 27).
Strong diffraction peaks were also visible for M16, revealing a high level of order in the colony, as supported by the structural analysis. As expected, such diffraction features are less prominent in M17 and absent in M5. By using the grating equation, the lattice constant for the different strains WT, M16, and M17 can be estimated as , , and , respectively (Materials and Methods and SI Appendix, Fig. S3). These values were consistent within the error margin of the SEM observations, considering the shrinkage that typically arises from fixation of the sample. The presence of strong diffraction peaks for WT and M16 reveals a high level of order in the colonies. In contrast, the lack of diffraction peaks in M5 indicates a poor organization of the bacteria (Fig. 2B), despite the presence of a certain degree of order in the autocorrelation (SI Appendix, Fig. S7F). This suggests that the packing of the M5 strain in the vertical direction is not well maintained (Fig. 3C) and that the bacteria are not forming distinct layers in the colony, as is the case for the other strains (Fig. 2B). M17, on the other hand, displays a weak and broad diffraction around and . In this case, even though the colony fails to create an ordered lattice within a layer (Fig. 2D), it probably maintains distinct layers in the vertical direction, thus enhancing coherent reflection.
We therefore conclude that each of the strains provides a specific optical fingerprint, which is attributed not only to the actual dimensions of the bacteria but also mainly to their ability to locally self-organize in a defined lattice. In fact, while the dimensions of WT and M5 were comparable, they displayed a radically different optical response. Moreover, M16 maintained correlation on a longer scale than the WT, indicating that it is possible to improve the regularity of the organization of living optical structures via genetic manipulation.
Analysis of Transposon Insertion Mutants Confirms Relation Between Motility and Structural Color
The ability of the bacteria to self-organize into a regular lattice is affected by cell-to-cell communication and motility as suggested by Kientz et al. (26). Therefore, we isolated mutants with defects in motility on ASWBC and ASWB Very Low nutrient (ASWBVLow) plates to explore this relationship.
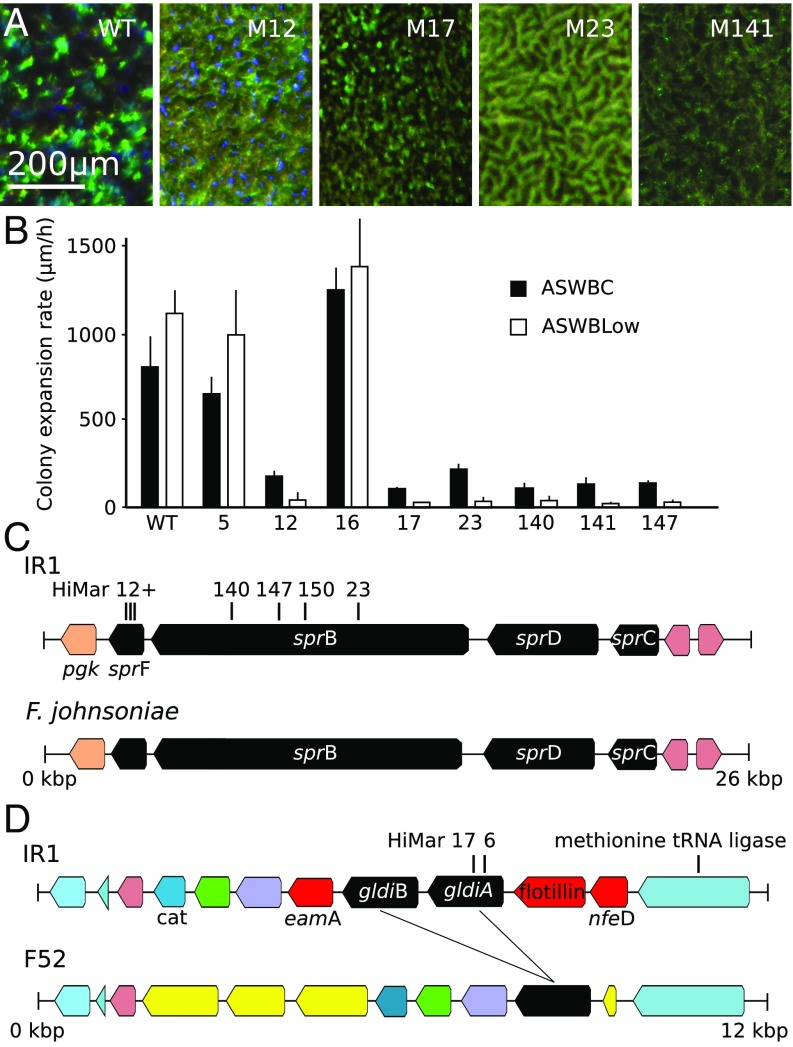
Fig. 4B reports the colony expansion rate (driven by motility but requiring growth) of different strains of the bacteria. By comparing the migration rate of WT and mutants obtained during screening of transposon libraries of IR1 on ASWBC plates, it is evident that gliding motility plays an important role in the organizational capability of the colonies compared with their visual appearance (Fig. 4 and SI Appendix, Fig. S8). It was notable that colonies displaying structural color showed a terraced appearance near the edge, suggestive of ordered layers. In particular, we observed that highly motile bacteria, like the WT and M16, organized on ASWBC plates in long-range periodic structures, while reduced-motility strains, such as M17, failed to do so. However, motility alone was insufficient to produce a significant order, as in the case of M5 (Fig. 2).
Fig. 4.
Phenotyping and mapping of transposon insertions affecting motility and iridescence. (A) Comparison of iridescence of WT IR1 and mutants viewed under the microscope and cultured on ASWBC agar overnight at 22°C. M12–M23 have been color enhanced since they would otherwise appear black in comparison. (B) The rate of colony expansion of the WT and mutant strains quantified on ASWBC and ASWVLow plates. (C) Mapping of transposon insertions of mutants 12, 23, 147, and 149 within the sprC–F gene cluster of strain IR1, showing close homology with a similar operon from F. johnsoniae UW101. (D) Mapping of mutants 6 and 17 within the gldiA gene of IR1 and comparison with the region of the Flavobacterium F52 genome that contains homologous genes (black, a single copy in F52 and two in IR1). Genes marked in yellow are found in this region only for F52. Genes in red (eamA, a flotillin motif-containing gene, and nfeD) are found in this region only in IR1. Other colors indicate ORFs found in this region in common between the two bacteria, including a putative methionine tRNA ligase upstream and the cat chloramphenicol resistance gene downstream.
Mutants with no detectable motility and dull coloration (M12, M23, M140, M147) were mapped to the sprB (sprB23::HiMar, sprB140::HiMar, sprB147::HiMar, sprB150::HiMar) and sprF (sprF12::HiMar and other isolates) genes; all located within the same gene cluster (Fig. 4C and SI Appendix, Fig. S2). In Flavobacterium johnsoniae, these spr genes formed part of a single operon with two other genes involved in gliding motility, gldC and gldD. This order of genes is conserved in IR1 (28). It is known that the SprB protein in F. johnsoniae is located on the cell surface and is required for motility on agar (29), while the SprF protein is involved in the assembly of SprB on the cell surface and infers a lack of motility of the bacteria on agar (28). Therefore, we conclude that the gliding motility in the IR1 has many core components similar to that of F. johnsoniae.
Mutants M6 and M17 have transposon insertions in different loci within the gldiA gene (gldiA6::HiMar and gldiA17::HiMar). This gene was the upstream copy of two closely related genes (gldiA and gldiB), encoding predicted polypeptides of with identity (Fig. 4D). The gldi genes were absent from the genome of F. johnsoniae UW101 (30). The gldi have no assigned function and are found only in a small number of bacteria, an exception being the rhizosphere Flavobacterium F52 (31). F52 single copy of a gldi gene ( and identity comparing the F52 gene to gldiA and gldiB, respectively) and there was a similar arrangement of these genes between IR1 and F52 (Fig. 4D). The genes immediately flanking gldiA and gldiB in IR1 were not conserved in F52 and were all related genes known to encode membrane or membrane-associated proteins, a member of the SPFH Flotillin group of proteins known to be involved in lipid binding and a member of the NFED membrane-anchored proteins (32) and an EamA family integral membrane protein (33). M6 and M17 also showed a dull coloration (e.g., Fig. 4A and SI Appendix, Fig. S8 J–L). Despite F52 not previously having been described as structurally colored, we observed colonies with structural color on ASWB and ASWBLow agar (SI Appendix, Fig. S8 C–F). The structural color of F52 was reduced in a nonspreading mutant with a knockout (KO) of the gldJ gene, which encodes the GldJ lipoprotein required for motility in Flavobacteria (34) (SI Appendix, Fig. S8).
In addition, another nonmotile and dull mutant of IR1 (M141) was also mapped to a homologue of the wzx gene (wzx141::HiMar, Fig. 4C), predicted to encode a MATE family protein. Close homologues to the IR1 wzx gene were found only in very few flavobacteria with no role described. However, this gene has been implicated in swarming motility mediated by flagella, facilitating wetting agar surfaces to promote migration (35). Taken together, it is clear that mutants with decreased motility affect the formation of structural color. The involvement of some of these motility genes could have been predicted from studies on other Flavobacteria (28–31) while the others reported here are unique or with a known function but not previously implicated in gliding motility.
Moreover, we observed that the WT structural coloration was lost by mixing the cells in a colony with a sterile inoculation loop but reformed rapidly within 5– (Movie S1), which required metabolic energy but not de novo protein synthesis (SI Appendix, Fig. S9). The motility mutants described above were similarly able to reform coloration, albeit more slowly ( to ) and creating a duller coloration than the WT strain, when replated on nutrient agars (SI Appendix, Fig. S8 G–L). However, unlike the WT, this rearrangement was not possible for nonmotile strains M12, M17, and M23 when transferred to agars nonpermissive for growth. It is therefore concluded that growth was not required for reforming colored structures in situations where gliding motility is active and the cell colony was previously structurally colored. The IR1 colony can therefore be considered a self-organizing system requiring only energy and a suitable surface to rapidly reform structural color.
We conclude that gliding motility is necessary to obtain efficient colony organization and observe a strong optical response. For mutants defective in gliding, cell growth and division may still facilitate sufficient cell movement and alignment to permit a limited degree of regular packing and color.
Not Only Motility Genes Affect Structural Coloration
In the genome of the dull but motile mutant M5, a single transposon insertion was found in a gene closely related to the spoT gene of Escherichia coli (SI Appendix, Fig. S2). The SpoT protein is an enzyme with the capability of degrading guanosine tetraphosphate (ppGpp), the alarmone that triggers a decrease in transcription of many genes (36). In E. coli a spoT KO leads to an elevated level of ppGpp. To test whether the activation of the stringent response (elevated level of ppGpp) inhibited iridescence, WT IR1 was grown with DL-serine hydroxamate (DLSH), which induces ppGpp synthesis via the inhibition of seryl-tRNA synthetase and the failure to charge the tRNA leading to starvation for that amino acid (37). DLSH was a strong inhibitor of iridescence, but not of gliding. The cells sustained rates of up to on ASWLow plates, which is similar to the WT. In other words, the effects of DLSH gave a similar phenotype to M5 (spoT::HiMar), inhibiting the formation of structural color. Two independent KOs (M49, M64) were obtained in a putative trmD tRNA methyltransferase ( identity). Such KOs completely abolished the structural coloration while allowing growth and motility, suggesting a possible role in the fidelity of translation in regulating structural color (38).
Moreover, possible interaction of the colonies with plants is suggested by the screening of three mutant strains, M40, M86, and M88. A transposon insertion (M40) into a putative GH3 family gene gave the phenotype of losing coloration faster than the WT (without dying faster). The GH3 family consists of enzymes known to structurally modify plant growth hormones, suggesting a link to the growth of IR1 on macroalgae; the latter are known to produce auxins, benzoates, and other hormones. The closest homology was to a GH3.12-containing polypeptide from Solanum lycopersicum (tomato), which modifies benzoates with a glutamine residue (39). While the predicted polypeptide was widely distributed in Flavobacteria, there is no evidence for the function in this family. In addition, a transposon insertion into a putative endoxylanase (M86 and M88) led to a dull phenotype on ASWBC agar.
Repeated independent transposition events resulted in the highly motile, highly organized colonies described in Figs. 1 and 2. Mutant 16 (hypA16::HiMar) was typical and analyzed in detail. The gene disrupted is one of unknown function, but predicted by domain motifs to encode a SRPBCC deep hydrophobic ligand-binding domain (40). Closely related genes to hypA were widely distributed in the Flavobacteria, for which automated annotation indicated a putative transcription factor. However, homology searches outside the Flavobacteria and domain searches failed to reveal any support for a role in transcription.
Presence of Macroalgae Influences the Growth of Bacteria
As structural coloration by Flavobacteria has been observed only on culture media but never in their natural environment, the biological significance of their coloration remains unclear (27). To investigate whether the periodic packing responsible for structural coloration can be sustained in a natural environment, we investigated the growth of IR1 strains on different surfaces available in estuarine environments and the effect of natural nutrients as well as in the presence of commonly available algal polymers.
It was found that fucoidan (a sulphated polymer from brown algae) and starch maintained the periodic packing of the WT and could tune the diffraction response toward colors at higher and lower wavelengths. Red, blue, and purple appearances were observed in addition to green when growing the WT on ASWBC. The purple/blue coloration was most obvious in young colonies and with high concentrations of starch or fucoidan above (wt/vol) (SI Appendix, Tables S1 and S2). After 5–8 d at C on most permissive nutrient agars, including ASWLow plates, coloration was completely lost. Addition of powdered fucoidan within 1–2 d of loss of structural coloration caused reordering of the cells of IR1 to allow recovery of structural color (Fig. 5B). These observations suggest that the presence of such nutrients is important in the natural environment of IR1 and that the loss of structural color in aging colonies is reversible (further screenings of natural surfaces are described in SI Appendix).
Fig. 5.
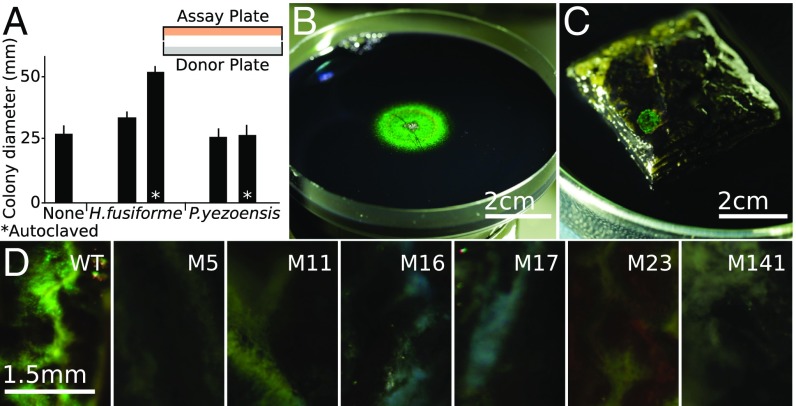
The effect of growth on algae and algal products directly and indirectly on IR1. (A) The effect of exposing IR1 to volatiles from the algae P. yezoensis and H. fusiforme. An ASWLow plate (assay plate) was inoculated in the center with a aliquot containing cfu of WT IR1. This was placed facing a second plate (donor plate) containing salt agar without algae or with of the specified hydrated algae (either autoclaved or not). Plates were sealed with parafilm and incubated for 7 d under humidified conditions. (B) Example of fucoidan-triggered recovery of structural color. The entire 9-cm diameter agar plate was spread with IR1, which became intensely green across the whole plate after 24 h and then lost coloration completely after 7 d. Addition of of powdered fucoidan to the center of the plate facilitated recovery of structural color after 1 d, as shown. (C) WT IR1 shown growing as a 6-mm diameter iridescent green colony on a - stack of P. yezoensis after at . The algae were embedded in agarose with (wt/vol) KCl and nigrosin. (D) Culture of WT and IR1 mutants on 4-mm diameter stalks of H. fusiforme after . (Scale bars in D, Left to Right, 1.5 mm.)
IR1 grown under most conditions failed to influence the growth or coloration of a second culture on another agar plate when separated by a 1-cm air gap from the second plate (Fig. 5A). Exposure to volatiles from IR1 cultured with fucoidan prolonged structural coloration for 3 d. This suggests that the bacteria were releasing influential volatiles from the breakdown of fucoidan. Volatiles from the red algae Pyropia yezoensis and brown algae Hijikia fusiforme were tested for their effects on the WT strain cultured on ASWB and ASWBVLow plates. Exposure to P. yezoensis had little effect, but on ASWB plates H. fusiforme stimulated growth and larger, more colored colonies. This effect was terminated either by ventilating the cultures or by autoclaving the algae (Fig. 5A). Taken together, these experiments suggested IR1 growing on algal products or exposed to brown algae could promote the organization of other cells of IR1 at a distance of at least .
The influence of algae and algal products, combined with the observations that IR1 formed structural color only on hydrated surfaces exposed to air and that the salinity optimum was 1% (wt/vol) suggested the possibility that structural color may form in nature in the estuarine environment. Biotic surfaces likely to be available to IR1 were screened for their ability to support growth of IR1 displaying structural color. Materials from both animals and macroalgae from estuarine environments were collected. By growing the WT on animal material (fish, crustacea) or rocks coated with microbial biofilms we were not able to observe any effect of structural coloration from the WT after 1–14 d. While organization on most macroalgae was limited, the surface of P. yezoensis derived from fresh or dried sources demonstrated it to be a good substrate with strong, green structural coloration forming rapidly after transfer with the greatest intensity after and persistence up to 2 d (Fig. 5C). The development of color was very similar in kinetics and appearance to ASWBC medium containing -carrageenan. Structural coloration ranging from green to blue was also observed on the surface of H. fusiforme (Fig. 5D). In this case, the coloration appeared after a few hours and persisted for up to 20 d, which is three times longer than on ASWBC medium where it generally lasts for only 1 wk.
Mutants were screened for their ability to form structurally colored colonies on H. fusiforme (Fig. 5D). The strains M5 (spoT5::HiMar) and M17 (gldiA17::HiMar) were impaired in coloration on ASWBC plates and did not form structurally colored colonies on the algal surface. M16 (hypA16::HiMar), despite being intensely colored on ASWBC agar, was not able to form colored colonies on the algae. Motility mutants M12 (sprF12::HiMar), M23 (sprB23::HiMar), and M141 (wsx141::HiMar) were able to form colored colonies only to a limited extent. These data suggest that the surfaces of red and particularly brown macroalgae (rich in fucoidan) can be good substrates for the organization of IR1 to form structurally colored colonies in nature and that motility assists organization on the surfaces.
Discussion
We studied the genotype and phenotype relations governing structural coloration in bacteria, showing the possibility to engineer structural color in a living organism. Flavobacterium IR1 showed a formidable capacity and flexibility to organize as a colony, drastically modifying its optical appearance in terms of spectral and angular response under different growth conditions and with genetic modification. We demonstrated that structural coloration of the colonies was linked to motility and other cellular functions including genes with no previously assigned function. Our observations shed light on the biological function of structural coloration in bacteria and suggest an interaction of IR1 with macroalgae. This may relate to photoprotection of bacteria or host or optimum organization to degrade biological polymers, with structural color a secondary consequence.
This presentation of IR1 as a rapidly self-organizing system, requiring only competent living cells sustaining a proton motive force and a suitable surface, showcases a highly flexible model organism, which can be used as a future biomaterial for photonic applications. As an example, dehydrated and fixed structures composed of bacteria can be engineered for paints and nanotemplates. Moreover, we envision viable pathways for engineering bacteria toward living sensors with intrinsic self-healing capabilities. As an example, they can be optimized for changing coloration under external stimuli and interface with other living tissues.
Materials and Methods
Details of the materials and methods are given in SI Appendix. Briefly, strain IR1 (a Flavobacterium isolated from Rotterdam Harbor) was grown on culture media with salinity matching the location where it was isolated (0.5–1.5%) or directly on the surface of macroalgae. The IR1 genome was sequenced by Illumina Hi-seq paired-end technology. More than 20,000 colonies from the HiMar transposon library were screened for mutants altered in color, with transposon insertions mapped on the genome as previously described (30). Colonies of IR1 were fixed under conditions which maintained structural color for electron microscopy. The arrangement of cells in colonies from SEM was measured in ImageJ. Autocorrelation analysis of the SEM images was furthermore performed using in-house computer scripts and is presented in SI Appendix. Optical analysis of structural color was by goniometry and was performed on live samples using a custom-made goniometer. Analysis of the recorded data relied on the grating equation and peak extraction was via in-house computer scripts. Additional data related to this publication are available at the University of Cambridge data repository (doi.org/10.17863/CAM.16794). The genome sequence of IR1 is available from GenBank under accession no. NQOT00000000.
Supplementary Material
Acknowledgments
C.J.I. thanks Philip de Groot and students and staff of Hogeschool Utrecht, Biobased Economy and Leiden for bioinformatics support and Marcel Giesbers for help with electron microscopy. C.J.I. thanks Mark McBride and Yongtao Zhu for assistance with transposon mutagenesis. V.E.J., S.V., and T.S.R. thank Lars Jelsbak for fruitful discussions. B.D.W. and S.V. thank Tobias Wenzel for discussions. V.E.J., M.M.S., and S.V. thank Beverley Glover for help with growing bacteria. C.J.I. thanks the Biotechnology-Based Ecologically Balanced Sustainable Industrial Consortium (BE-Basic) Foundation (The Netherlands) for financial support. V.E.J., S.V., and T.S.R. thank the Biotechnology and Biological Sciences Research Council (BBSRC) David Phillips fellowship (BB/K014617/1), the European Research Council (ERC-2014-STG H2020 639088), and the European Commission [Marie Curie Fellowship Looking Through Disorder (LODIS), 701455] for financial support. B.D.W. was financially supported through the National Center of Competence in Research Bio-Inspired Materials and the Ambizione program of the Swiss National Science Foundation (168223).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. NQOT00000000). Additional data related to this publication are available at the University of Cambridge data repository (doi.org/10.17863/CAM.16794).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716214115/-/DCSupplemental.
References
- 1.Kinoshita S. Structural Colors in the Realm of Nature. World Scientific; Toh Tuck, Singapore: 2008. [Google Scholar]
- 2.Kinoshita S, Yoshioka S. Structural colors in nature: The role of regularity and irregularity in the structure. ChemPhysChem. 2005;6:1442–1459. doi: 10.1002/cphc.200500007. [DOI] [PubMed] [Google Scholar]
- 3.Vukusic P, Sambles JR. Photonic structures in biology. Nature. 2003;424:852–855. doi: 10.1038/nature01941. [DOI] [PubMed] [Google Scholar]
- 4.Prum RO, Torres RH. A fourier tool for the analysis of coherent light scattering by bio-optical nanostructures. Integr Comp Biol. 2003;43:591–602. doi: 10.1093/icb/43.4.591. [DOI] [PubMed] [Google Scholar]
- 5.Wiersma DS. Disordered photonics. Nat Photon. 2013;7:188–196. [Google Scholar]
- 6.Vukusic P, Hallam B, Noyes J. Brilliant whiteness in ultrathin beetle scales. Science. 2007;315:348. doi: 10.1126/science.1134666. [DOI] [PubMed] [Google Scholar]
- 7.Chandler CJ, Wilts BD, Brodie J, Vignolini S. Structural color in marine algae. Adv Opt Mater. 2017;5:1600646. [Google Scholar]
- 8.Hahnke RL, Harder J. Phylogenetic diversity of Flavobacteria isolated from the north sea on solid media. Syst Appl Microbiol. 2013;36:497–504. doi: 10.1016/j.syapm.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Kientz B, Agogué H, Lavergne C, Marié P, Rosenfeld E. Isolation and distribution of iridescent Cellulophaga and other iridescent marine bacteria from the charente-maritime coast, French Atlantic. Syst Appl Microbiol. 2013;36:244–251. doi: 10.1016/j.syapm.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Doucet SM, Meadows MG. Iridescence: A functional perspective. J R Soc Interface. 2009;6:S115–S132. doi: 10.1098/rsif.2008.0395.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilts BD, Michielsen K, De Raedt H, Stavenga DG. Sparkling feather reflections of a bird-of-paradise explained by finite-difference time-domain modeling. Proc Natl Acad Sci USA. 2014;111:4363–4368. doi: 10.1073/pnas.1323611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi NN, et al. Keeping cool: Enhanced optical reflection and radiative heat dissipation in Saharan silver ants. Science. 2015;349:298–301. doi: 10.1126/science.aab3564. [DOI] [PubMed] [Google Scholar]
- 13.Li L, et al. A highly conspicuous mineralized composite photonic architecture in the translucent shell of the blue-rayed limpet. Nat Commun. 2015;6:6322. doi: 10.1038/ncomms7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs M, et al. Photonic multilayer structure of Begonia chloroplasts enhances photosynthetic efficiency. Nat Plants. 2016;2:16162. doi: 10.1038/nplants.2016.162. [DOI] [PubMed] [Google Scholar]
- 15.Whitney HM, et al. Floral iridescence, produced by diffractive optics, acts as a cue for animal pollinators. Science. 2009;323:130–133. doi: 10.1126/science.1166256. [DOI] [PubMed] [Google Scholar]
- 16.Whitney HM, Reed A, Rands SA, Chittka L, Glover BJ. Flower iridescence increases object detection in the insect visual system without compromising object identity. Curr Biol. 2016;26:802–808. doi: 10.1016/j.cub.2016.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghiradella HT, Butler MW. Many variations on a few themes: A broader look at development of iridescent scales (and feathers) J R Soc Interface. 2009;6:S243–S251. doi: 10.1098/rsif.2008.0372.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prum RO, Dufresne ER, Quinn T, Waters K. Development of colour-producing -keratin nanostructures in avian feather barbs. J R Soc Interface. 2009;6:S253–S265. doi: 10.1098/rsif.2008.0466.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Onelli OD, et al. Development of structural colour in leaf beetles. Sci Rep. 2017;7:1373. doi: 10.1038/s41598-017-01496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilts BD, et al. Butterfly gyroid nanostructures as a time-frozen glimpse of intracellular membrane development. Sci Adv. 2017;3:e1603119. doi: 10.1126/sciadv.1603119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuthill IC, et al. The biology of color. Science. 2017;357:eaan0221. doi: 10.1126/science.aan0221. [DOI] [PubMed] [Google Scholar]
- 22.Shrivastava A, Lele PP, Berg HC. A rotary motor drives Flavobacterium gliding. Curr Biol. 2015;25:338–341. doi: 10.1016/j.cub.2014.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McBride MJ. Bacterial gliding motility: Multiple mechanisms for cell movement over surfaces. Annu Rev Microbiol. 2001;55:49–75. doi: 10.1146/annurev.micro.55.1.49. [DOI] [PubMed] [Google Scholar]
- 24.Cousin S, Päuker O, Stackebrandt E. Flavobacterium aquidurense sp. nov. and Flavobacterium hercynium sp. nov., from a hard-water creek. Int J Syst Evol Microbiol. 2007;57:243–249. doi: 10.1099/ijs.0.64556-0. [DOI] [PubMed] [Google Scholar]
- 25.Bernardet JF, Nakagawa Y, Holmes B. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol. 2002;52:1049–1070. doi: 10.1099/00207713-52-3-1049. [DOI] [PubMed] [Google Scholar]
- 26.Kientz B, et al. Glitter-like iridescence within the bacteroidetes especially Cellulophaga spp.: Optical properties and correlation with gliding motility. PLoS One. 2012;7:1–12. doi: 10.1371/journal.pone.0052900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kientz B, et al. A unique self-organization of bacterial sub-communities creates iridescence in cellulophaga lytica colony biofilms. Sci Rep. 2016;6:19906. doi: 10.1038/srep19906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhodes RG, Nelson SS, Pochiraju S, McBride MJ. Flavobacterium johnsoniae sprB is part of an operon spanning the additional gliding motility genes sprC, sprD, and sprF. J Bacteriol. 2011;193:599–610. doi: 10.1128/JB.01203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson SS, Bollampalli S, McBride MJ. Sprb is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J Bacteriol. 2008;190:2851–2857. doi: 10.1128/JB.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McBride MJ, et al. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl Environ Microbiol. 2009;75:6864–6875. doi: 10.1128/AEM.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolton M, et al. Draft genome sequence of Flavobacterium sp. strain f52, isolated from the rhizosphere of bell pepper (Capsicum annuum l. cv. maccabi) J Bacteriol. 2012;194:5462–5463. doi: 10.1128/JB.01249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hinderhofer M, et al. Evolution of prokaryotic SPFH proteins. BMC Evol Biol. 2009;9:10. doi: 10.1186/1471-2148-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Västermark Å, Almén MS, Simmen MW, Fredriksson R, Schiöth HB. Functional specialization in nucleotide sugar transporters occurred through differentiation of the gene cluster eama (duf6) before the radiation of Viridiplantae. BMC Evol Biol. 2011;11:123. doi: 10.1186/1471-2148-11-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolton M, Frenkel O, Elad Y, Cytryn E. Potential role of flavobacterial gliding-motility and type IX secretion system complex in root colonization and plant defense. MPMI. 2014;27:1005–1013. doi: 10.1094/MPMI-03-14-0067-R. [DOI] [PubMed] [Google Scholar]
- 35.Toguchi A, Siano M, Burkart M, Harshey RM. Genetics of swarming motility in Salmonella enterica serovar typhimurium: Critical role for lipopolysaccharide. J Bacteriol. 2000;182:6308–6321. doi: 10.1128/jb.182.22.6308-6321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laffler T, Gallant J. spot, a new genetic locus involved in the stringent response in E. coli. Cell. 1974;1:27–30. [Google Scholar]
- 37.Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. Rele, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci USA. 2001;98:14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hori H. Methylated nucleosides in tRNA and tRNA methyltransferases. Front Genet. 2014;5:1–26. doi: 10.3389/fgene.2014.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westfall CS, Herrmann J, Chen Q, Wang S, Jez JM. Modulating plant hormones by enzyme action: The gh3 family of acyl acid amido synthetases. Plant Signal Behav. 2010;5:1607–1612. doi: 10.4161/psb.5.12.13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iyer LM, Koonin EV, Aravind L. Adaptations of the helix-grip fold for ligand binding and catalysis in the start domain superfamily. Proteins Struct Funct Genet. 2001;43:134–144. doi: 10.1002/1097-0134(20010501)43:2<134::aid-prot1025>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.