Significance
Flowering in many plants begins with the perception of daylength in leaves. For example, some plants flower in short days, others in long days. In Arabidopsis thaliana, the leaves then transmit a small protein, FLOWERING TIME T (FT), to the shoot apex in the food-conducting cells, the phloem. Arrival of FT causes the apex to transition from leaf to flower formation. In this paper, we show that only two files of phloem cells in A. thaliana veins synthesize FT. This is also true for the Maryland Mammoth cultivar of tobacco (Nicotiana tabacum). Killing the cells specifically affects downstream, but not upstream, genes, indicating that an extensive intercellular signaling system regulates FT synthesis in the phloem.
Keywords: flowering, phloem, minor veins, signaling, diphtheria toxin
Abstract
Flowering is triggered by the transmission of a mobile protein, FLOWERING LOCUS T (FT), from leaves to the shoot apex. FT originates in the phloem of leaf veins. However, the identity of the FT-synthesizing cells in the phloem is not known. As a result, it has not been possible to determine whether the complex regulatory networks that control FT synthesis involve intercellular communication, as is the case in many aspects of plant development. We demonstrate here that FT in Arabidopsis thaliana and FT orthologs in Maryland Mammoth tobacco (Nicotiana tabacum) are produced in two unique files of phloem companion cells. These FT-activating cells, visualized by fluorescent proteins, also activate the GALACTINOL SYNTHASE (CmGAS1) promoter from melon (Cucumis melo). Ablating the cells by expression of the diphtheria toxin gene driven by the CmGAS1 promoter delays flowering in both Arabidopsis and Maryland Mammoth tobacco. In Arabidopsis, toxin expression reduces expression of FT and flowering-associated genes downstream, but not upstream, of FT. Our results indicate that specific companion cells mediate the essential flowering function. Since the identified cells are present in the minor veins of two unrelated dicotyledonous species, this may be a widespread phenomenon.
Flowering in Arabidopsis thaliana and in many other species is induced by the synthesis of a small, mobile protein FLOWERING LOCUS T (FT) in leaves (1–3), followed by transmission to the shoot apex (4). FT transcription is regulated by a complex system of converging upstream regulatory pathways (4–6). Although FT synthesis is known to occur in the phloem (7), the specific site(s) at which this takes place has not been determined due to diffusion of the commonly used GUS reporter gene product (8) which typically produces blue coloration over the entire vein. This is especially problematic in Arabidopsis studies because the veins are complex (9). As a result, the intercellular dynamics of FT synthesis have not been resolved.
To increase resolution, we turned to fluorescent proteins, clearing the tissue with ClearSee (10), which removes chlorophyll but retains fluorescent protein reporters. Our results indicate that FT synthesis is confined to two noncontiguous files of phloem companion cells that appear in some, but not all, transverse sections of the vein as a pair. Unexpectedly, the same cells activate the GALACTINOL SYNTHASE1 (GAS1) gene from melon (11), the first dedicated gene in the raffinose-oligosaccharide (RFO) pathway. Ablation of these cells by expression of the diphtheria toxin gene driven by the CmGAS1 promoter affects the expression of downstream flowering genes, but has a limited effect on the genes that control FT synthesis. Similar ablation experiments on the Maryland Mammoth (MM) cultivar of tobacco also interfere with flowering. These results indicate that FT synthesis is restricted to specific companion cells in the veins of Arabidopsis and tobacco, unrelated long- and short-day plants, respectively, and suggest that flowering regulation in these plants, and presumably others, involves previously unrecognized patterns of intercellular signaling.
Results
Identification of FT-Synthesizing Cells in Arabidopsis.
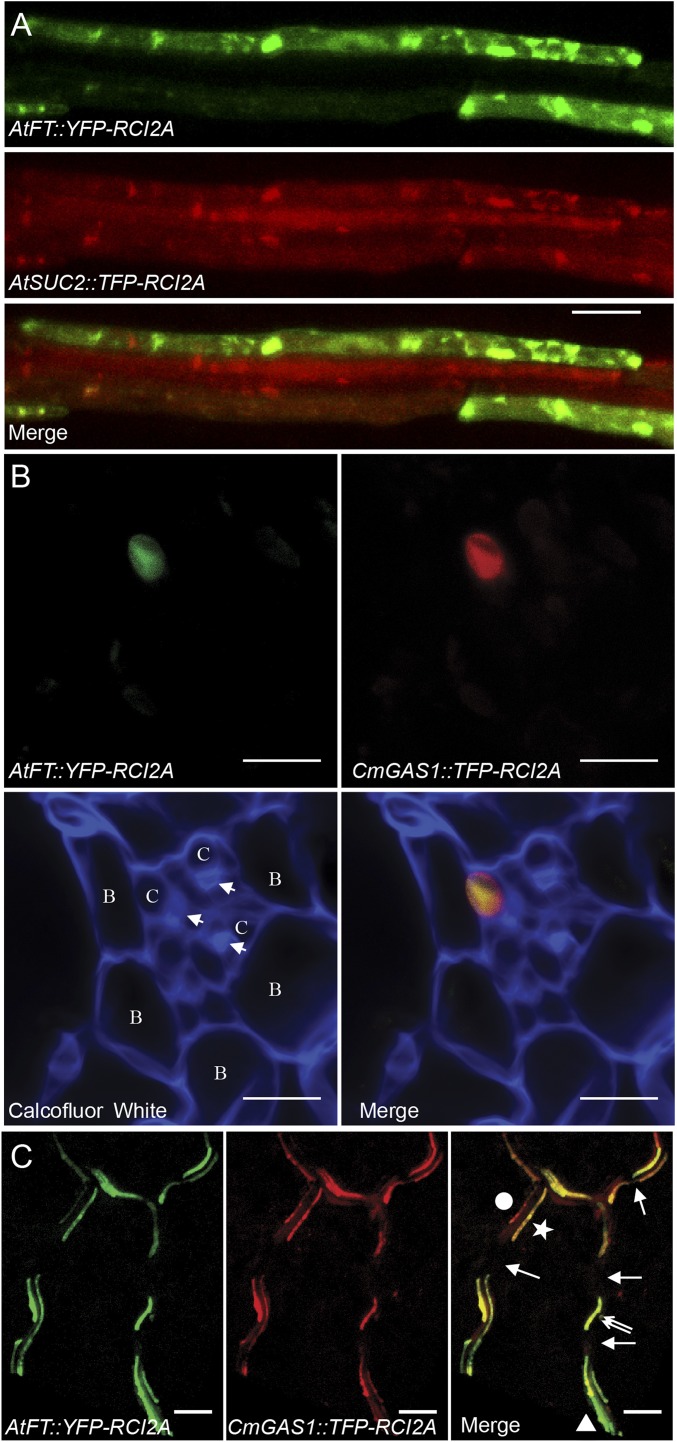
To determine the site of FT synthesis in Arabidopsis, we transformed plants with a construct in which the AtFT promoter drives the gene encoding yellow fluorescent protein (YFP) (Fig. 1). Certain of these plants were also transformed with the gene for the teal fluorescent protein (TFP) driven by the ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 2 (AtSUC2) promoter, which is specific to companion cells (12) (Fig. 1A). In both cases, the fluorescent probes were anchored to membranes by the hydrophobic amino acid sequence RCI2A (13) to prevent cell-to-cell movement. Viewed from the upper side of the leaf, cells activating AtFT appear as two files on opposite sides of a vein (Fig. 1A). These cells also activate the AtSUC2 promoter, indicating that they are a subset of companion cells. This is consistent with the observation that flowering is induced when AtFT is driven by the AtSUC2 promoter (14). Note that the files of companion cells with AtFT activity are discontinuous; not all companion cells in the file activate the FT gene.
Fig. 1.
Localization of promoter activities in Arabidopsis veins by confocal microscopy. (A) Paradermal view from the top of the leaf. Cells that activate the AtFT promoter (AtFT::YFP-RCI2A) and the companion cell-specific AtSUC2 promoter (AtSUC2::TFP-RCI2A) fluoresce green and red, respectively. (B) Transverse hand section of a minor vein. Cell walls are made visible at the bottom by Calcofluor white staining (blue fluorescence). Fluorescence from the AtFT (green) and CmGAS1 (red) promoters overlap in a single companion cell. B, bundle sheath cell; C, companion cell. The arrowhead indicates a sieve element. (C) Low-magnification, paradermal views of veins from the upper side of the leaf illustrating superimposition of AtFT (green) and CmGAS1 (red) promoter activities. Each vein is apparent as two adjacent, parallel files of cells. The circle and star in the merged image are placed on opposite sides of a single vein, in which two files of companion cells run parallel to each other. Note that the cell closest to the dot activates the CmGAS1 promoter more strongly that the AtFT promoter. The companion cell indicated by the triangle in the bottom right of the image activates the AtFT promoter more strongly than the CmGAS1 promoter. In all veins, gaps in staining are evident, indicating weak or no promoter activity in either of the companion cells (arrows). In some locations in a vein, there is little if any staining in one of the two companion cells (double arrows), as is also seen in B. Tissues in A and C were cleared with ClearSee. (Scale bars: 10 µm in A and B; 50 µm in C.)
The AtFT gene expression pattern in specific companion cells on opposite sides of leaf veins is similar to that of the GALACTINOL SYNTHASE1 (CmGAS1) gene from melon (11). Galactinol synthesis is the first step in raffinose-family oligosaccharide (RFO) synthesis in the cucurbits and a limited number of other families that phloem load by polymer trapping. Interestingly, The CmGAS1 promoter is also active in Arabidopsis, tobacco, and gray poplar (Populus tremula X alba), which do not phloem load by the polymer trap mechanism (15).
To determine whether FT is synthesized in these cells, we cotransformed Arabidopsis plants with AtFT::YFP-RCI2A and CmGAS1::TFP-RCI2A. In the transverse (cross-) section of a vein from such a plant, signals driven by the two promoters overlap in one of the cells in the adaxial region of the vein (Fig. 1 B). Note that in the lower-magnification images in Fig. 1C, relative signal intensities generated by the AtFT and CmGAS1 promoters often differ in individual cells and that, as also seen in Fig. 1A, the signals are discontinuous along the length of the veins, resulting in gaps between fluorescent cells (arrows in Fig. 1C). Additional confocal images are shown in Figs. S1 and S2.
Since vein topography differs in juvenile and adult leaves, we prepared tissue from cotyledons and from mature, true leaves of different plant ages before flowering. The same pattern of twin cells activating both the CmGAS1 and AtFT promoters was found in all samples.
Ablation of CmGAS1-Expressing Cells Delays Flowering.
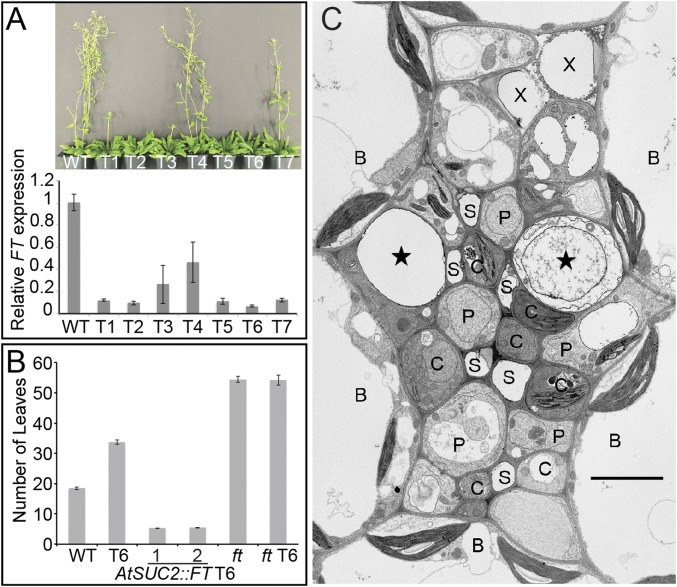
To determine whether CmGAS1-expressing cells control flowering in Arabidopsis, we used the CmGAS1 promoter to drive expression of the diphtheria toxin A gene (DT-A), a translation inhibitor that induces cell death (16). Plants from multiple independent lines grew normally, but in inductive, long-day conditions (16 h light, 8 h dark), expression of FT was significantly diminished (Fig. 2A) and flowering was significantly delayed (Fig. 2B and Fig. S3), although not to the same degree as in ft mutants. Fig. 2C is an electron micrograph of a vein from toxin-expressing line T6, with severely delayed flowering. Two cells in the adaxial region of the vein, abutting the bundle sheath, are essentially devoid of content. The rest of the phloem appears uninjured. The earlier flowering in toxin-expressing plants compared with ft mutants (Fig. 2B) could be due to weak activity of the CmGAS1 promoter in some cells (Fig. 1C). Although it is possible that FT is also produced in another cell type, no such signal from the AtFT::YFP-RCI2A gene construct was identified by confocal analysis of tissues from more than 200 leaves. In noninductive, short days (10 h light, 14 h dark), wild-type (WT) and transgenic plants flowered at approximately the same time and stage of development (Fig. S4), indicating that the flowering result in long days is not due to general debilitation of the plants by the toxin.
Fig. 2.
Plants expressing the diphtheria toxin gene CmGAS1::DT-A. (A) Phenotype of WT and transgenic (T1–T7) plants expressing the diphtheria toxin gene CmGAS1::DT-A. (Top) Representative plants grown in long days (16 h day/8 h night) photographed on day 55. (Bottom) Relative FT expression levels in different CmGAS1::DT-A transgenic lines compared with WT. Error bars represent SE. n = 3. (B) Leaf numbers were scored when plants flowered. CmGAS1::DT-A transgenic line T6 flowers later than WT. Driving expression of FT with the AtSUC2 promoter accelerates flowering in plants with ablated CmGAS1-activating cells. Two representative independent lines, AtSUC2::FT-1 and AtSUC2::FT-2, in a T6 background were used. ft mutants with ablated CmGAS1-activating cells (ft T6) flower at the same time as ft mutants. (C) Electron micrograph of a vein in an A. thaliana plant transformed with the DT-A gene driven by the CmGAS1 promoter. Note the destruction of cytoplasm in two companion cells (stars) adjacent to bundle sheath (B) cells. This is a larger vein than that shown in Fig. 1B. C, additional companion cells; P, phloem parenchyma cells; S, sieve elements; X, xylem elements. Error bars represent SE. n = 3 in A; n ≥ 15 in B. (Scale bar: 4 µm in C.)
To confirm that delayed flowering in plants expressing CmGAS1::DT-A toxin is due to a reduction of FT mRNA, we transformed toxin line T6 with AtSUC2::FT. Most of the transformed lines flowered much earlier than WT, at approximately the 5-leaf stage (Fig. 2B and Fig. S5A). To further confirm that diminished FT expression causes the flowering defect in toxin lines, we crossed T6 and the ft mutant. The progeny flowered at the same time as the ft mutants (Fig. 2B and Fig. S5B), indicating that the ft mutation is epistatic to the toxin effect and late flowering in the toxin-expressing lines is due to reduced FT expression.
Gene Expression in CmGAS1-Activating Cells.
To identify expressed genes enriched in CmGAS1-activating cells, we compared expression in WT and CmGAS1::DT-A–expressing plants by RNA sequencing analysis (RNA-seq). Leaves were collected at 8 h and 16 h after lights on from plants grown in flower-inducing, long-day conditions; 196 genes were up-regulated and 84 genes were down-regulated at 8 h, and 379 genes were up-regulated and 60 genes were down-regulated at 16 h (Dataset S1). Since many of the up-regulated genes are defense-related, we assume that they were activated in response to the toxin. We also make the simplifying assumption that ablation results in reduced expression of genes normally active in these cells. Therefore, we restricted further analyses to genes down-regulated in CmGAS1::DT-A–expressing plants. We combined the genes down-regulated at 8 h (76 genes), 16 h (52 genes), or both (8 genes) (Fig. S6A) and conducted a Gene Ontology (GO) analysis on the list by agriGO (17). Six GO terms were significantly enriched in our gene list (Fig. S6 B and C). Among these, three are related to flowering, confirming that regulation of flowering is a major function of cells that activate the CmGAS1 promoter.
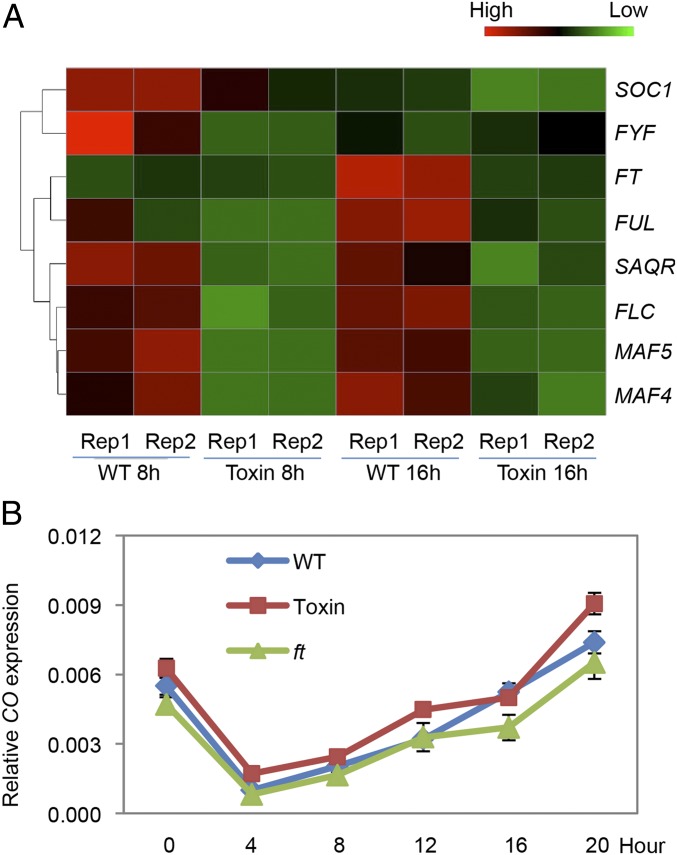
Among the 136 down-regulated genes, expression of FT was strongly reduced, as expected (Fig. 3A). All well-known genes upstream of FT in the photoperiodic pathway were unaffected by CmGAS1::DT-A, including CONSTANS (Fig. 3B and Fig. S6D). Seven genes known to regulate flowering, in addition to FT, were down-regulated by CmGAS1::DT-A (Fig. 3A). FRUITFUL (FUL), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), and FOREVER YOUNG FLOWER (FYF) act downstream of FT (18–20). Since FT is transported from leaves to the shoot apex, we cannot conclude that down-regulation of these genes occurs in CmGAS1-activating cells. FLOWERING LOCUS C (FLC), MADS AFFECTING FLOWERING 4 (MAF4), and MADS AFFECTING FLOWERING 5 (MAF5) are in a clade of homologs that negatively regulate flowering (21). The expression of these genes is not altered in ft mutants, but expression of all three was strongly reduced by the diphtheria toxin (Fig. 3A and Fig. S7). The strongest reductions of gene expression by CmGAS1::DT-A were in FT and MAF5. Reduced expression in a family of genes that negatively regulates FT suggests that a fine-tuning mechanism exists in these cells for controlling the release of flowering signals. The SAQR (SENESCENCE-ASSOCIATED AND QQS-RELATED; AT1G64360) gene (22, 23), which is only slightly down-regulated in the ft mutants (P = 0.097), was also down-regulated in CmGAS1::DT-A–expressing plants (Fig. 3A and Fig. S7).
Fig. 3.
The toxin-expressing cell is a platform for flowering genes. (A) Heatmap plot generated from RNA sequencing. Rep1 and Rep2 are replicates. Toxin indicates CmGAS1::DT-A. The expression of eight flowering-related genes is significantly decreased by the toxin. (B) CONSTANS transcript abundance is not decreased by toxin. Expression level is relative to actin. Error bars represent SE. n = 3.
Gene Expression in MM Tobacco.
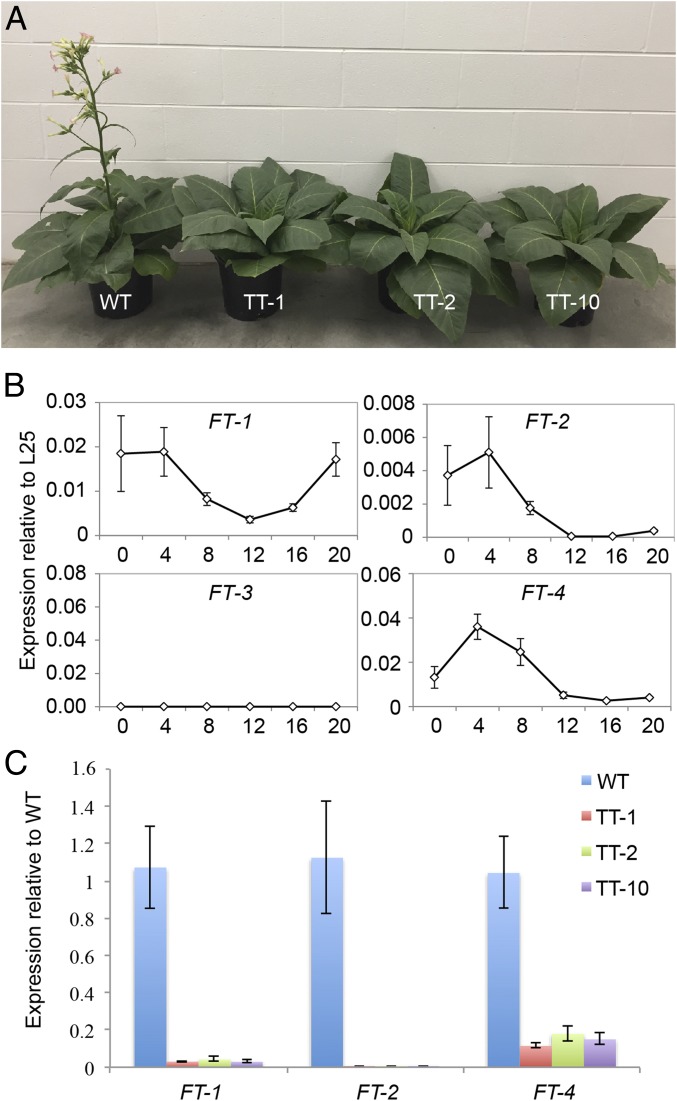
To determine if the flowering signal is produced in specific companion cells in leaves of other species, we turned to the MM tobacco cultivar (Fig. 4). MM tobacco, which flowers in short days, was one of the plants in which photoperiodicity in the flowering response was discovered (24). Of eight independent transgenic lines grown in short days (8 h light, 16 h dark), three died within 2 mo, one flowered by 100 d (as did WT), two (TT2 and TT7) flowered by 150 d, and the remaining two lines, TT1 and TT10, grew to >2 m in height over the next 8 mo without flowering. The leaves of these plants were normal in size and appearance (Fig. 4A). When the TT10 shoot was grafted to a flowering WT MM stem, it produced flowers after 40 d, demonstrating that the shoot apex was capable of flowering.
Fig. 4.
CmGAS1 promoter activity in tobacco. (A) MM tobacco WT and toxin lines TT-1, TT-2, and TT-10 grown for 8 wk in short days. (B) Diurnal relative expression profiles of the four FT-like genes in WT MM tobacco (L25 as a reference gene). FT-3 expression is below quantifiable limits. (C) Fold change in expression of FT-like genes in MM tobacco transgenic lines TT-1, TT-2, and TT-10 compared with WT. Bars in the PCR plots represent SEs. n = 4.
In day-neutral tobacco leaves (Nicotiana tabacum cv. SR1), four FT-like genes are expressed in short days (25). NtFT1–3 are floral inhibitors, while NtFT4 is a floral inducer. In our experiments on MM tobacco grown in short days (8 h light, 16 h dark), expression of NtFT1, 2, and 4 peaked at ZT 4 h, while NtFT3 expression was below quantifiable limits (Fig. 4B). Measured at 4 h into the day period, NtFT1, NtFT2, and NtFT4 mRNA levels were all reduced in the three CmGAS1::DA-T lines with delayed flowering (TT1, TT2, and TT10), from 5- to 33-fold (Fig. 4C), indicating that transcription of AtFT orthologs in tobacco takes place in, or is controlled by, cells that activate the CmGAS1 promoter.
Discussion
It has been known for longer than a decade that the flowering regulator, FT (6, 7), and at least some additional components of the flowering pathways, including the immediate upstream activator CONSTANS (26), are synthesized in the phloem of leaf veins. However, the complexity and somewhat irregular arrangement of cells in veins has hindered more detailed analysis. The phloem of leaf minor veins is composed of sieve elements, companion cells, phloem parenchyma cells, and, in some cases, thick-walled cells with protective functions (27). Parenchyma cells and companion cells may be of similar size, again making identification problematic. Another significant difficulty is presented by the positioning of veins in the middle of the lamina, surrounded by heavily pigmented mesophyll and bundle sheath cells. This pigmentation interferes with observation of fluorescent reporter proteins, which may explain previous reliance on the GUS reporter system. However, while GUS is useful in localizing gene expression at the tissue level, diffusion of the primary reaction product between cells (28) obscures the initial site of synthesis.
To improve resolution, we took advantage of the ClearSee clearing agent to view fluorescent reporter proteins within the veins. We also identified cells that activate the CmGAS1 gene by using the promoter to drive expression of the diphtheria toxin gene, which specifically kills cells in which the toxin is synthesized. By this combination of techniques, the site of AtFT activity was revealed to be in cells that activate melon GALACTINOL SYNTHASE (CmGAS1), even though Arabidopsis is not a polymer trap plant. Activity of the CmGAS1 promoter in cells that do not phloem load by the polymer trap mechanism suggests that the CmGAS1 promoter responds to a conserved regulatory mechanism with function beyond that of RFO synthesis, but only in specific companion cells of the leaf.
Although we cannot definitively conclude based on microscopic analysis that the spatial activities of the AtFT and CmGAS1 promoters are identical, they clearly overlap to a considerable degree. The reason for this coexpression pattern is not obvious, but one possibility is that, in an evolutionary sense, the RFO pathway has co-opted the flowering regulatory network. This would explain why the CmGAS1 promoter is active in the FT-synthesizing cells. Nevertheless, the genes seem to be not closely coregulated, except for cell specificity. Relative activities of the two genes often differ, and previous results have indicated that CONSTANS, an immediate upstream regulator of FT expression in Arabidopsis, is not needed for CmGAS1 activation (29).
In addition, it should be noted that both CmGAS1 (11) and AtFT (figure 4A in ref. 5) promoters are activated first in the leaf tip and then progressively toward the base as the leaf ages. This is the common basipetal pattern of leaf development in dicotyledonous plants (30). Basipetal development explains why FT expression is often evident only in leaf tips when young plants, with only partially mature leaves, are used in promoter studies.
Given that Arabidopsis and MM tobacco are members of the two largest clades of dicotyledonous plants (the rosids and asterids, respectively), restriction of FT transcription to two cell files in veins may be a common pattern. Why is FT synthesized in specific companion cells? One possible reason is that these cells are specialized in some way for intercellular communication. Indeed, their common positioning next to the bundle sheath on the two sides of the vein could facilitate interaction with the photosynthetic tissue. However, other companion cells in the veins are also capable of transmitting the flowering signal, since plants in which the GAS1-activating cells are destroyed by the diphtheria toxin flower early when transformed with AtSUC2::FT. Another possibility, which is not incompatible with one above, is that flowering regulation is too complex to function properly if all interacting upstream factors are present together in the FT-synthesizing cells. Perhaps restriction of FT synthesis to specific cells in the veins makes possible nuances of gene interaction with other members of the flowering pathway over space and time.
Materials and Methods
Plant Material and Growth Condition.
Seeds of A. thaliana Columbia-0 (Col-0) were soaked in water for 2 d at 4 °C and then sown in soil mix. Plants were propagated under long days (16 h light, 8 h dark) or short days (10 h light, 14 h dark), maintained at 22 °C with 60 µmol m−2 s−1. Tobacco was grown under short days (8 h light, 16 h dark) and maintained at 25 °C with 300 µmol m−2 s−1.
Cloning and Transformation.
Two primers, pUC19-GASF and pUC19-GASR, were used to amplify the fragment of the CmGAS1 promoter from the genomic DNA of melon (C. melo) (11). pUC19 was digested with XbaI and SacI and then fused with the CmGAS1 promoter digested with the same enzymes to form a new vector, pUC-GAS. DT-A toxin was amplified with pUC-ToxinF and pUC-ToxinR from DT-A plasmid (GenBank accession no. AB535096), and then digested with KpnI and SacI and fused to the KpnI and SacI sites of pUC-GAS digested with the same enzymes to form a vector, pUC-GAS-Toxin. The GAS-Toxin fragment was released from pUC-GAS-Toxin by digestion with XbaI and SacI and then ligated to the pGPTV vector to make pGPTV-GAS-Toxin for plant transformation.
The CmGAS1 promoter was cloned into a plant expression vector, pER8, using the Gibson assembly method with the PCR product of the primers 8xhoGASF and 8speGASR to get pER8-GASp. The YFP gene fused with RCI2A (AT3G05880) genomic DNA for membrane targeting (a gift from the A. Roeder laboratory, Cornell University, Ithaca, NY) was cloned into the SpeI site of pER8-GASp to make pER8-GASp:YFP-RCI2A. The CmGAS1 promoter was also cloned into plant expression vector pORE-O2 to make pO2-GASp. TFP-RCI2A was cloned between the PstI and SpeI sites of pO2-GASp to make pO2-GASp:TFP-RCI2A.
The 2,237-bp promoter of the AtSUC2 gene and TFP-RCI2A gene (a gift from the A. Roeder laboratory, Cornell University, Ithaca, NY) were assembled together to the pORE-O2 vector using four primers—o2suc2pxhoF, suc2pTFP, TFPsuc2p, and o2TFPRCIpstR—through the Gibson assembly cloning method to make pO2-SUC2p:TFP-RCI2A.
The 8,095-bp promoter of the FT gene was cloned into pER8 using the Gibson assembly method with the PCR product of two primers—8FTpxhoF and 8FTpxhoR—to make pER8-FTp. YFP-RCI2A was then cloned into the XhoI site of pER8-FTp to make pER8-FTp:YFP-RCI2A.
pGPTV-GAS-Toxin, pER8-GASp:YFP-RCI2A, pO2-GASp:TFP-RCI2A, pO2-SUC2p:TFP-RCI2A, and pER8-FTp:YFP-RCI2A were transformed into Col-0 mediated by Agrobacterium tumefaciens GV3101 strain using the floral spray method. For fluorescent colocalization analysis, transgenic plants with pER8-GASp:YFP-RCI2A and pO2-SUC2p:TFP-RCI2A, pER8-FTp:YFP-RCI2A and pO2-SUC2p:TFP-RCI2A, or pER8-FTp:YFP-RCI2A and pO2-GASp:TFP-RCI2A were crossed, and confocal imaging was performed on F1 plants. pGPTV-GAS-Toxin was transformed into MM tobacco as described previously (11).
Microscopy.
The ClearSee method (10) was used to localize fluorescence generated by CmGAS1::TFP-RCI2A and AtFT::YFP-RCI2A or CmGAS1::YFP-RCI2A and AtSUC2::TFP-RCI2A in minor veins of adult leaves. Leaves were fixed in Pipes buffer (70 mM, pH 6.8) with 4% paraformaldehyde and 1% glutaraldehyde for 2 h at room temperature and washed in Pipes buffer (70 mM, pH 6.8) three times, and then incubated in ClearSee solution for 1–4 wk at room temperature. Fluorescence signals were also collected from fresh leaf tissue prepared by hand sectioning with a razor blade. In some preparations, cell walls were stained with 1 mg/mL Calcofluor white for 1 min. Confocal imaging was perform with a Zeiss LSM 710 microscope using a smart design setting for YFP, TFP or Calcofluor white. The paradermal views of the vein were built on Z-series of images by Z-stack imaging the entire vein. Gains for the individual fluorophores were adjusted for optimal intensity; similarities in intensity for different constructs do not indicate similar promoter activities. The images were processed in ImageJ (31).
RNA Extraction and Quantitative PCR.
RNA was extracted using the Plant RNA Extraction Kit (Sigma-Aldrich) in accordance with the manufacturer’s protocol with slight modifications. DNase treatment was performed using on-column DNase (Sigma-Aldrich) in accordance with the manufacture’s recommendation. cDNA was synthesized using 2 µg of total RNA, 100 pmol Oligo dT, 10 nmol dNTP, 20 U of RiboLock RNase inhibitor (Thermo Fisher Scientific), and 200 U of RevertAid reverse transcriptase (Thermo Fisher Scientific) in a 20-µL reaction. Quantitative PCR was performed in 96-well plates on a Bio-Rad iQ5 machine using iTaq universal SYBR Green Supermix (Bio-Rad) in a 12-µL reaction. Gene expression was estimated based on threshold cycles (Ct) normalized to the expression of housekeeping genes. Primers used for gene expression studies are listed in Table S1.
RNA Library Preparation and RNA-Seq.
An Illumina sequencing library was prepared as described previously (32). In brief, polyadinylated RNA was separated from 5 µg of total RNA using Dynabeads oligo(dT)25 (Invitrogen) and then fragmented. The first- and second-strand cDNAs were generated using dNTP and dUTP, respectively. After end-repair, dA-tailing and TruSeq Y-shaped adapter ligation, the dUTP-containing second strand was digested. The resulting library was PCR-amplified with TruSeq-indexed PCR primers and sequenced on the Illumina HiSeq4000 platform at the Core Genomics Facility of Weill Cornell Medical College.
The RNA-seq analysis protocol was adopted a previously described protocol (33). The reads were collected and verified by sequence quality control with ShortRead. Using Tophat2 software, the reads were aligned to the reference genome Arabidopsis_thaliana.TAIR10.30.gtf, which was downloaded from plants.ensembl.org/info/website/ftp/index.html. The bam files from Tophat2 were sorted by name through Samtools software and used to generate count files by htseq-count software. A differential expression analysis was then conducted with the edgeR package with default parameters and two replicates under a simple design strategy. The genes were selected as differentially expressed with a 15% false discovery rate and are listed in Dataset S1, in which tair_locus is the gene ID, logFC is logarithm of fold-changes in gene expression, logCPM is logarithm of counts per million reads, P Value is P value, FDR is false discovery rate, and tair symbol is known gene name. Heatmap plots were generated by the pheatmap package in R.
Electron Microscopy.
Leaf tissue was fixed in 2% (vol/vol) glutaraldehyde plus 2% (vol/vol) paraformaldehyde in 70 mM Pipes buffer, pH 6.8, for 1 h at room temperature, then washed and postfixed in 1% (vol/vol) osmium tetroxide in the same buffer. The tissue was dehydrated in an ethanol series and embedded in Spurr’s epoxy resin (Electron Microscopy Sciences). Thin sections were stained with uranyl acetate and lead citrate and observed with a Philips EM-300 transmission electron microscope.
Supplementary Material
Acknowledgments
We thank A. Roeder (Cornell University) for the use of the confocal microscope and for technical advice, and A. Jagendorf (recently deceased) for helpful discussions. The paper was read critically by A. Jagendorf, T. Owens, B. G. Turgeon, and H. Turgeon. Electron microscopy was conducted by Richard Medville of Electron Microscopy Services. This work was supported by National Science Foundation Grant IOS-1354718 (to R.T.) and startup funds from Purdue University (to C.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719455115/-/DCSupplemental.
References
- 1.Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 2.Lin MK, et al. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell. 2007;19:1488–1506. doi: 10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- 4.Zeevaart JAD. Leaf-produced floral signals. Curr Opin Plant Biol. 2008;11:541–547. doi: 10.1016/j.pbi.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Shim JS, Kubota A, Imaizumi T. Circadian clock and photoperiod flowering in Arabidopsis: Constans is a hub for signal integration. Plant Physiol. 2017;173:5–15. doi: 10.1104/pp.16.01327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otero S, Helariutta Y. Companion cells: A diamond in the rough. J Exp Bot. 2017;68:71–78. doi: 10.1093/jxb/erw392. [DOI] [PubMed] [Google Scholar]
- 7.Takada S, Goto K. Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell. 2003;15:2856–2865. doi: 10.1105/tpc.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Ruijter NCA, et al. Evaluation and comparison of the GUS, LUC and GFP reporter system for gene expression studies in plants. Plant Biol. 2003;5:103–115. [Google Scholar]
- 9.Haritatos E, Medville R, Turgeon R. Minor vein structure and sugar transport in Arabidopsis thaliana. Planta. 2000;211:105–111. doi: 10.1007/s004250000268. [DOI] [PubMed] [Google Scholar]
- 10.Kurihara D, Mizuta Y, Sato Y, Higashiyama T. ClearSee: A rapid optical clearing reagent for whole-plant fluorescence imaging. Development. 2015;142:4168–4179. doi: 10.1242/dev.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haritatos E, Ayre BG, Turgeon R. Identification of phloem involved in assimilate loading in leaves by the activity of the galactinol synthase promoter. Plant Physiol. 2000;123:929–937. doi: 10.1104/pp.123.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Truernit E, Sauer N. The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of beta-glucuronidase to the phloem: Evidence for phloem loading and unloading by SUC2. Planta. 1995;196:564–570. doi: 10.1007/BF00203657. [DOI] [PubMed] [Google Scholar]
- 13.Thompson MV, Wolniak SM. A plasma membrane-anchored fluorescent protein fusion illuminates sieve element plasma membranes in Arabidopsis and tobacco. Plant Physiol. 2008;146:1599–1610. doi: 10.1104/pp.107.113274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbesier L, Coupland G. Photoperiodic flowering of Arabidopsis: Integrating genetic and physiological approaches to characterization of the floral stimulus. Plant Cell Environ. 2005;28:54–66. [Google Scholar]
- 15.Slewinski TL, Zhang CK, Turgeon R. Structural and functional heterogeneity in phloem loading and transport. Front Plant Sci. 2013;4:244. doi: 10.3389/fpls.2013.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collier RJ. Effect of diphtheria toxin on protein synthesis: Inactivation of one of the transfer factors. J Mol Biol. 1967;25:83–98. doi: 10.1016/0022-2836(67)90280-x. [DOI] [PubMed] [Google Scholar]
- 17.Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teper-Bamnolker P, Samach A. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell. 2005;17:2661–2675. doi: 10.1105/tpc.105.035766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoo SK, et al. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 2005;139:770–778. doi: 10.1104/pp.105.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duplat-Bermúdez L, Ruiz-Medrano R, Landsman D, Mariño-Ramírez L, Xoconostle-Cázares B. Transcriptomic analysis of Arabidopsis overexpressing flowering locus T driven by a meristem-specific promoter that induces early flowering. Gene. 2016;587:120–131. doi: 10.1016/j.gene.2016.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y. Control of the transition to flowering by chromatin modifications. Mol Plant. 2009;2:554–564. doi: 10.1093/mp/ssp005. [DOI] [PubMed] [Google Scholar]
- 22.Jones DC, et al. A clade-specific Arabidopsis gene connects primary metabolism and senescence. Front Plant Sci. 2016;7:983. doi: 10.3389/fpls.2016.00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luhua S, Ciftci-Yilmaz S, Harper J, Cushman J, Mittler R. Enhanced tolerance to oxidative stress in transgenic Arabidopsis plants expressing proteins of unknown function. Plant Physiol. 2008;148:280–292. doi: 10.1104/pp.108.124875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garner WW, Allard HA. Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. J Agric Res. 1920;18:553–606. [Google Scholar]
- 25.Harig L, et al. Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J. 2012;72:908–921. doi: 10.1111/j.1365-313X.2012.05125.x. [DOI] [PubMed] [Google Scholar]
- 26.An H, et al. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development. 2004;131:3615–3626. doi: 10.1242/dev.01231. [DOI] [PubMed] [Google Scholar]
- 27.Sack L, Scoffoni C. Leaf venation: Structure, function, development, evolution, ecology and applications in the past, present and future. New Phytol. 2013;198:983–1000. doi: 10.1111/nph.12253. [DOI] [PubMed] [Google Scholar]
- 28.Jefferson RA. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol Biol Report. 1987;5:387–405. [Google Scholar]
- 29.Ayre BG, Turgeon R. Graft transmission of a floral stimulant derived from CONSTANS. Plant Physiol. 2004;135:2271–2278. doi: 10.1104/pp.104.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turgeon R. The sink-source transition in leaves. Annu Rev Plant Physiol. 1989;40:119–138. [Google Scholar]
- 31.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong S, et al. High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc. 2011;2011:940–949. doi: 10.1101/pdb.prot5652. [DOI] [PubMed] [Google Scholar]
- 33.Anders S, et al. Count-based differential expression analysis of RNA sequencing data using R and bioconductor. Nat Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.