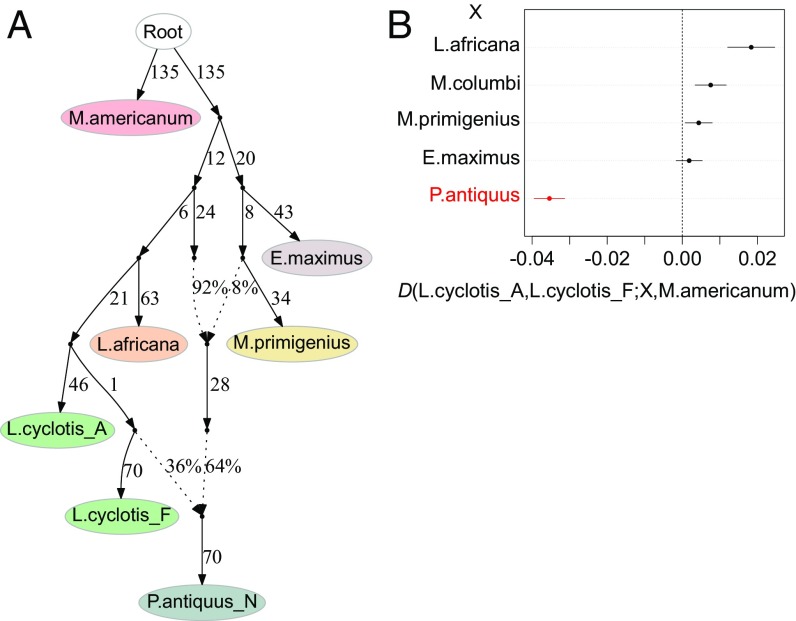
Fig. 2.
Admixture graph of elephantid history and supporting D-statistics. (A) Model of the phylogenetic relationships among elephantids augmented with admixture events. Branch lengths are given in drift units × 1,000. Two admixture events are inferred in the history of the straight-tusked elephant lineage, from a population related to woolly mammoths and a population related to the West African forest elephant (L. cyclotis_F) while most of its ancestry derives from a lineage most closely related to the common ancestor of savanna and forest elephants. We were not able to resolve the order of the two admixture events. Inferred ancestry proportions are ∼6 to 10% and 35 to 39% (with confidence intervals including uncertainty due to possible reference biases) (SI Appendix, Figs. S12.3 and S12.4) for the woolly mammoth-related and forest elephant-related components, respectively. (B) D-statistics testing for asymmetric genetic affinity between each of the two forest elephants and another elephantid (X). Positive values indicate excess genetic affinity between L. cyclotis_A and X while negative values indicate excess genetic affinity between L. cyclotis_F and X. Bars correspond to one SE in either direction. The statistic highlighted in red is significant (|Z| > 3) and indicates an excess of shared derived alleles between the straight-tusked elephant and L. cyclotis_F. Remaining key D-statistics supporting the admixture graph are shown in Table 2. All inferences are based on transversion polymorphisms only.