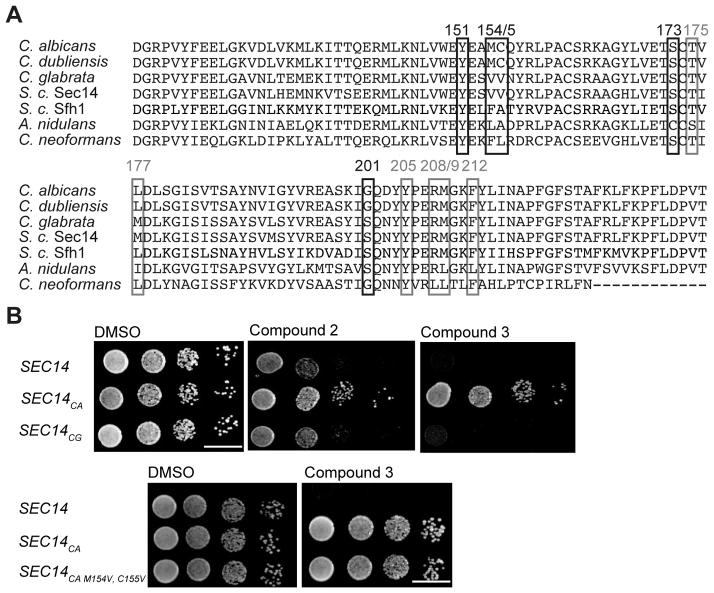
Figure 5.
Sec14p sequence comparison and investigations into the VV motif. A) Protein sequence alignment of Saccharomyces cerevisiae Sec14p and Sfh1p and Sec14p of the pathogenic fungi Candida albicans, Candida dubliniensis, Candida glabrata, Aspergillus nidulans and Cryptococcus neoformans. Resistance-conferring amino acids identified by the functional variomics screen are marked with black boxes, sites predicted by co-crystal structure to be involved in compound interactions with grey boxes. B) A wildtype strain and isogenic derivatives expressing physiological levels of C. albicans and C. glabrata Sec14 PITPs were spotted on rich medium (YPD) agar medium supplemented with the indicated compounds and incubated at 30 °C for 48 hours. Transplacement of the VV-motif into Sec14pCA (the M154V, C155V double mutant) did not render this PITP sensitive to compound 3 (lower panel).