SUMMARY
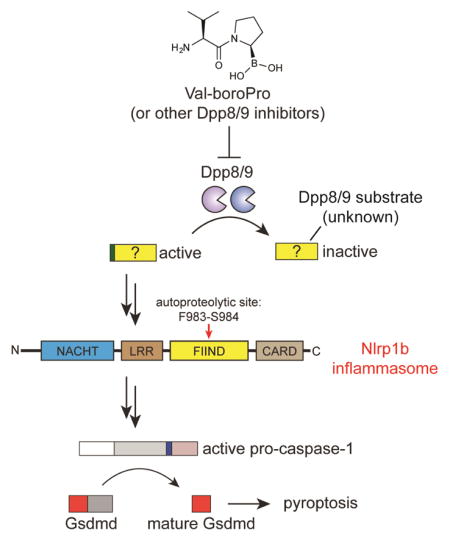
Val-boroPro (PT-100, Talabostat) induces powerful anti-tumor immune responses in syngeneic cancer models, but its mechanism of action has not yet been established. Val-boroPro is a non-selective inhibitor of post-proline-cleaving serine proteases, and the inhibition of the highly related cytosolic serine proteases Dpp8 and Dpp9 (Dpp8/9) by Val-boroPro was recently demonstrated to trigger an immunostimulatory form of programmed cell death known as pyroptosis selectively in monocytes and macrophages. Here we show that Dpp8/9 inhibition activates the inflammasome sensor protein Nlrp1b, which in turn activates pro-caspase-1 to mediate pyroptosis. This work reveals a previously unrecognized mechanism for activating an innate immune pattern recognition receptor, and suggests that Dpp8/9 serve as an intracellular checkpoint to restrain Nlrp1b and the innate immune system.
eTOC blurb
Dpp8/9 inhibitors induce an inflammatory form of cell death called pyroptosis in monocytes and macrophages. Here, Okondo et al. show that Dpp8/9 inhibition activates the inflammasome sensor protein Nlrp1b, which in turn activates pro-caspase-1 to mediate pyroptosis. This work reveals a previously unrecognized mechanism for stimulating the innate immune system.
INTRODUCTION
Val-boroPro (Fig. 1A) is a nonselective inhibitor of post-proline cleaving serine proteases that induces immune-mediated tumor regressions in multiple mouse models of cancer (Adams, et al., 2004; Walsh, et al., 2013). Val-boroPro dramatically increases the mouse serum protein levels of several immunomodulatory cytokines, including G-CSF and CXCL1/KC, and it has been speculated that these cytokines mediate the antitumor responses (Adams, et al., 2004; Walsh, et al., 2013). However, the mechanistic basis for the production of these cytokines has remained enigmatic since their discovery in the early 2000s.
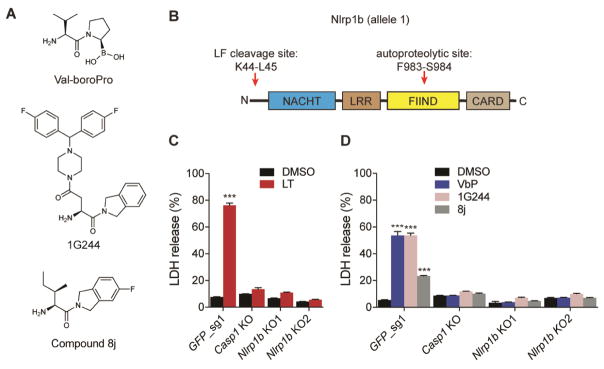
Figure 1. Nlrp1b is required for Dpp8/9 inhibitor-induced pyroptosis. See also Figure S1.
(A) Structures of Val-boroPro, 1G244, and compound 8j. (B) Diagram of mouse Nlrp1b allele 1. The LF cleavage and FIIND autoproteolysis sites are indicated. (C,D) Casp1 and Nlrp1b KO RAW 264.7 macrophages are resistant to LT (1 μg/mL, 6 h) (C) and the Dpp8/9 inhibitors Val-boroPro (2 μM, 24 h), 1G244 (5 μM, 24 h), and compound 8j (20 μM, 24 h) (D). Data are means ± SEM of three biological replicates. ***p < 0.001 by two-sided Student’s t-test for vehicle versus treated cells. LT, lethal toxin; VbP, Val-boroPro.
We recently discovered that Val-boroPro induces a lytic form of programmed cell death known as pyroptosis selectively in monocytes and macrophages (Okondo, et al., 2017; Taabazuing, et al., 2017). Briefly, the inhibition of the cytosolic serine dipeptidases Dpp8 and Dpp9 (Dpp8/9) by Val-boroPro, as well as by more selective Dpp8/9 inhibitors, activates the pro-protein form of the cysteine protease caspase-1 without autoproteolysis. This form of active pro-caspase-1 then cleaves gasdermin D (GSDMD), releasing its N-terminal region to form pores in the plasma membrane and mediate lytic cell death (Kayagaki, et al., 2015; Shi, et al., 2015). Consistent with the importance of this pathway to the immunostimulatory effects of Val-boroPro, Val-boroPro does not induce cytokines in mice lacking caspase-1 (Okondo, et al., 2017). However, it remains entirely unknown how Dpp8/9 inhibition results in the activation of pro-caspase-1.
Caspase-1-dependent pyroptosis, often referred to as “canonical pyroptosis”, is a critically important part of the innate immune response to pathogens(Broz and Dixit, 2016; Lamkanfi and Dixit, 2014). Canonical pyroptosis is typically triggered by microbial structures and activities, such as anthrax lethal toxin (LT), bacterial flagellin, and double stranded DNA, which are collectively referred to as pathogen-associated molecular patterns (PAMPs). When a PAMP enters the cytosol of a host cell, a specific sensor protein called a pattern-recognition receptor (PRR) either directly or indirectly detects its presence. PRRs then oligomerize with the adapter ASC to form large multiprotein signaling platforms called inflammasomes, which then recruit and trigger the proximity-induced autoproteolytic maturation and activation of caspase-1. Currently, five PRRs are known to form inflammasomes: Nlrp1, Nlrc4, Nlrp3, Aim2, and Pyrin. We hypothesized that Dpp8/9 inhibitors might activate one of these five PRRs through an unknown mechanism. Notably, Nlrp1 and Nlrc4 both contain caspase activation and recruitment domains (CARDs) that can directly interact with the CARD domain of pro-caspase-1 (Poyet, et al., 2001), and, in the absence of ASC, these two PRRs can activate the pro-protein form of caspase-1 without inflammasome formation or autoproteolysis (Broz, et al., 2010; Guey, et al., 2014; Van Opdenbosch, et al., 2014). As Dpp8/9 inhibitors similarly induce a form of pro-caspase-1-dependent pyroptosis independent of ASC (Okondo, et al., 2017), we speculated that either Nlrp1 or Nlrc4 might be mediating this pyroptotic response. Here we show that inhibition of Dpp8/9 activates the Nlrp1 inflammasome.
RESULTS
Nlrp1b is required for Dpp8/9 inhibitor-induced pyroptosis
We first wanted to investigate whether Nlrp1 was involved, as, unlike Nlrc4, the mechanisms controlling its activation remain poorly understood. The murine genome encodes three Nlrp1 paralogs, Nlrp1a, Nlrp1b, and Nlrp1c, but only Nlrp1b is known to form an inflammasome in response to pathogenic stimuli (Boyden and Dietrich, 2006; Ewald, et al., 2014; Gorfu, et al., 2014). Nlrp1b contains nucleotide-binding (NACHT), leucine-rich repeat (LRR), “function-to-find” (FIIND), and CARD domains (Fig. 1B) (Boyden and Dietrich, 2006), and undergoes post-translational autoproteolysis within its FIIND domain to generate N- and C-terminal fragments that remain associated in an auto-inhibited state (D’Osualdo, et al., 2011; Finger, et al., 2012; Frew, et al., 2012). Anthrax lethal toxin (LT) is the best-characterized activator of Nlrp1b (Boyden and Dietrich, 2006). LT is a bipartite toxin consisting of lethal factor (LF), a zinc metalloprotease, and protective antigen (PA), a cell binding-protein that transports LF into the host cell cytosol. Once cytosolic, LF directly cleaves Nlrp1b near its N-terminus (Fig. 1B), which is thought to relieve intramolecular auto-inhibition or induce a conformational change that enables inflammasome formation (Chavarria-Smith and Vance, 2013; Hellmich, et al., 2012; Levinsohn, et al., 2012). We first generated mouse RAW 264.7 macrophages lacking Nlrp1b (Fig. S1A), and confirmed that they, like Casp1−/− macrophages, are resistant to LT (Fig. 1C). Notably, we also found that Nlrp1b−/− RAW 264.7 macrophages were resistant to Val-boroPro (Fig. 1D) and the structurally unrelated Dpp8/9 inhibitors 1G244 (Jiaang, et al., 2005) and compound 8j (Van Goethem, et al., 2008) (Fig. 1A,D). In contrast, RAW 264.7 macrophages lacking Nlrc4 remained sensitive to Val-boroPro (Fig. S1B,C). These data indicate that Nlrp1b is required for Dpp8/9 inhibitor-induced pyroptosis.
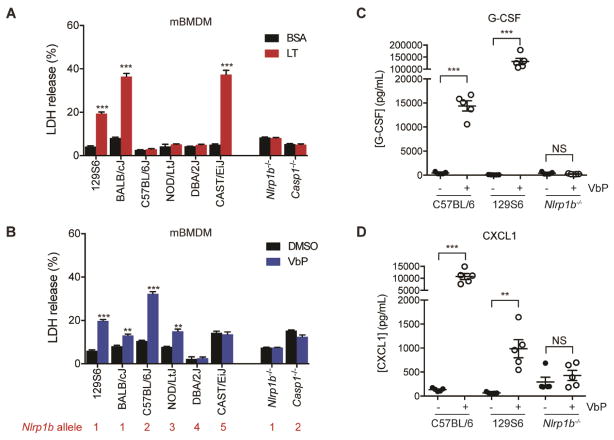
We next wanted to confirm that Nlrp1b is similarly required for Dpp8/9 inhibitor-induced pyroptosis in primary mouse macrophages and for cytokine induction in vivo. Nlrp1 is an extraordinarily polymorphic protein, with considerable variation in primary sequence both within and across species (Boyden and Dietrich, 2006; Newman, et al., 2010; Zhong, et al., 2016). Common inbred mouse strains have 5 different Nlrp1b alleles, and LT induces rapid pyroptosis only in macrophages carrying Nlrp1b alleles 1 or 5 (Fig. 2A, Fig. S2A, RAW 264.7 cells were derived from BALB/c mice and therefore have allele 1) (Boyden and Dietrich, 2006). Given the variable sensitivity of macrophages to LT, we first tested the responsiveness of macrophages from a panel of inbred mouse strains to Dpp8/9 inhibitors. Unlike LT, we found that macrophages containing alleles 1, 2, and 3, but not alleles 4 and 5 were sensitive to Val-boroPro (Fig. 2B, Fig. S2A) and compound 8j (Fig. S2B). Nlrp1b allele 4 contains a frameshift that generates a premature stop codon before the essential CARD domain (Boyden and Dietrich, 2006; Finger, et al., 2012; Frew, et al., 2012; Zhong, et al., 2016), and consequently these macrophages do not have a functional Nlrp1b protein. Therefore, the resistance of DBA/2J macrophages to Val-boroPro is consistent with a requirement for Nlrp1b in the Dpp8/9 inhibitor-induced pyroptotic pathway. CAST/EiJ mice, which are the only strain containing Nlrp1b allele 5, are genetically distant from the other inbred species and differ in a number of complex phenotypes (Davis, et al., 2007). It is not currently clear why these macrophages are resistant to Dpp8/9 inhibitors even though they express a functional Nlrp1b, but this is intriguing and warrants further study. We next obtained Nlrp1b−/− mice from The Jackson Laboratory. These knockout mice were generated on a 129S6 mouse background (Kovarova, et al., 2012), which contain a functional Nlrp1b allele but nonfunctional Nlrp1a and Nlrp1c alleles (Boyden and Dietrich, 2006), and therefore are entirely Nlrp1-deficient. Since their generation, these Nlrp1b−/− mice have been bred to C57BL/6J mice for at least six generations, but have been confirmed to retain complete Nlrp1 deficiency. We found that Nlrp1b−/− macrophages were resistant to LT (Fig. 2A), Val-boroPro (Fig. 2B), and compound 8j (Fig. S2B). Moreover, Val-boroPro induced serum G-CSF (Fig. 2C) and CXCL1 (Fig. 2D) in control 129S6 and C57BL/6J mice, but not in Nlrp1b−/− or DBA/2J mice (Fig. 2C,D, Fig. S2C–E). Thus, Nlrp1b is required for Val-boroPro-induced pyroptosis in primary mouse macrophages and for the induction of cytokines in vivo.
Figure 2. Nlrp1b controls primary mouse macrophages sensitivity to Val-boroPro. See also Figure S2.
(A,B) Primary macrophages from the indicated inbred mouse strains were treated with LT (1 μg/mL, 6 h) (A) or Val-boroPro (10 μM, 24 h) (B) before LDH release was assessed. The Nlrp1b allele encoded in each macrophage is shown below the strain in red (see also Fig. S2A). Data are means ± SEM of three biological replicates. ** p < 0.01, ***p < 0.001 by two-sided Student’s t-test for vehicle versus treated cells. (C,D) Val-boroPro (100 μg/mouse) induces high levels of serum G-CSF (C) and CXCL1/KC (D) after 6 h in C57BL/6 and 129S6 ice, but not in Nlrp1b−/−, mice as measured by ELISA. Data are means ± SEM, n = 5 mice/group. **p < 0.01, ***p < 0.001 by two-sided Student’s t-test for vehicle versus Val-boroPro-treated mice. NS, not significant.
Nlrp1b activation requires FIIND domain autoproteolysis
We next wanted to further investigate the molecular details of Nlrp1b activation by Val-boroPro. LT activation of Nlrp1b requires FIIND domain autoproteolysis (Frew, et al., 2012). To determine if autoproteolysis was similarly necessary for Val-boroPro-induced pyroptosis, we nucleofected plasmids encoding either wild-type or autoproteolysis-deficient S984A mutant Nlrp1b (allele 1) into Nlrp1b−/− RAW 264.7 macrophages. We found that only the wild-type protein restored sensitivity to LT and Val-boroPro (Fig. 3A, Fig. S3), demonstrating that FIIND domain autoproteolysis is indeed required for pyroptosis induced by Val-boroPro as well.
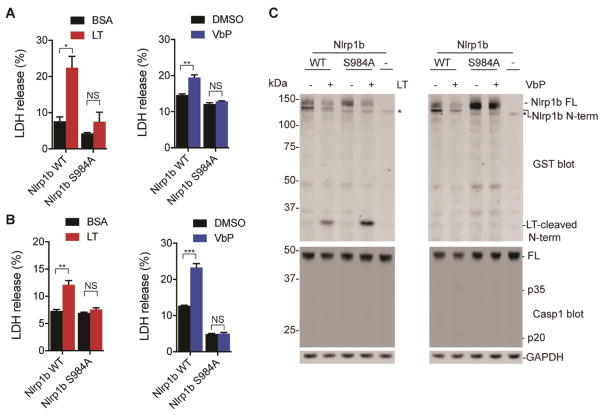
Figure 3. Activation of Nlrp1b requires FIIND autoproteolysis. See also Figure S3.
(A) Plasmids encoding WT or autoproteolysis-deficient mutant (S984A) Nlrp1b were nucleofected into Nlrp1b−/− RAW 264.7 cells, which were then treated with LT (1 μg/mL, 6 h) or Val-boroPro (10 μM, 24 h). Cytotoxicity was assessed by LDH release. (B,C) HEK 293T cells ectopically expressing mCasp1 and either Nlrp1b WT or S984A were treated with LT (1 μg/mL, 6 h) or Val-boroPro (5 μM, 24 h). Cytotoxicity was assessed by LDH release in (B) and Nlrp1b N-terminal cleavage was assessed by immunoblotting (construct contains an N-terminal GST tag) in (C). Asterisks indicate background bands. FL, full-length. In (A) and (B), data are means ± SEM of three biological replicates. *p<0.05, **p < 0.01, ***p < 0.001 by two-sided Student’s t-test for vehicle versus compound-treated cells. NS, not significant.
Ectopic expression of procaspase-1 and Nlrp1b is sufficient to confer sensitivity to LT in HT-1080 (Liao and Mogridge, 2009) and HEK293T (Chavarria-Smith, et al., 2016) cells, and we next wondered if recombinant expression of these components would similarly sensitize HEK 293T cells to Val-boroPro. Indeed, we found that expression of procaspase-1 and wild-type Nlrp1b also sensitized HEK 293T cells to Val-boroPro (Fig. 3B), demonstrating that these proteins are the only essential components of the Dpp8/9 pyroptotic pathway missing from these non-myeloid cells. In contrast, co-expression of procaspase-1 and S984A Nlrp1b did not confer sensitivity to HEK 293T cells, confirming that autoproteolysis is required for Nlrp1b activity. Moreover, Val-boroPro did not induce any observable N-terminal cleavage of Nlrp1b, indicating that Val-boroPro, unlike LT, does not activate Nlrp1b by direct proteolysis (Fig. 3C). As expected, very little, if any, caspase-1 cleavage was observed in response to either agent.
Dpp8/9 inhibitors require proteasome activity to induce pyroptosis
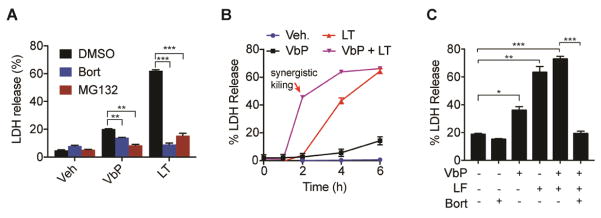
Interestingly, proteasome activity is required for macrophage sensitivity to LT (Fink, et al., 2008; Squires, et al., 2007; Tang and Leppla, 1999), although the mechanistic basis for this requirement is not understood. We wondered if Val-boroPro’s activation also required proteasome activity, and therefore treated RAW 264.7 cells with the proteasome inhibitors MG132 and bortezomib before the addition of Val-boroPro. Similar to LT, both MG132 and bortezomib blocked cell death induced by Val-boroPro (Fig. 4A). It should be noted that total amount of LDH release by Val-boroPro is lower than usual here as the LDH was assessed 6 h instead of 24 h to avoid apoptosis induced by proteasome inhibitors over longer intervals. Because LT and VbP activate Nlrp1b via different mechanisms, we hypothesized that these stimuli might induce synergistic cell death if administered simultaneously. Indeed, we observed that LT and VbP are remarkably synergistic (Fig. 4B), inducing ~50% LDH release at a time point (2 h) when either agent alone had little to no effect. Intriguingly, bortezomib completely blocks this synergistic cell death (Fig. 4C), highlighting the necessity of proteasome activity for the activation of Nrlp1b.
Figure 4. Proteasome inhibitors block Nlrp1b activation.
RAW 264.7 cells were treated with VbP (2 μM), LT (1 μg/mL), or both for the 6 h (a and c) or indicated time (b). In (a) and (c), cells were pretreated bortezomib (100 °C) or MG132 (100 °C) for 30 min prior to the addition of VbP and/or LT. Cytotoxicity was assessed by LDH release. In (b), LT and VbP induce synergistic cell death. Data are means ± SEM of three biological replicates. *p < 0.05, **p < 0.01, ***p < 0.001 by two-sided Students t-test.
DISCUSSION
In summary, we have now demonstrated that inhibition of Dpp8/9 activates pyroptosis via the Nlrp1b inflammasome. We found that Dpp8/9 inhibitors and LT require both Nlrp1b FIIND domain autoproteolysis and proteasome activity to trigger pyroptosis, but that Dpp8/9 inhibitors and LT activate different subsets of Nlrp1b alleles and only LT activates Nlrp1b by direct cleavage. Thus, the mechanisms of Nlrp1b activation by LT and Dpp8/9 inhibitors share some similarities, but are notably distinct.
Projecting forward, many important aspects of the Dpp8/9 inhibitor-induced pyroptotic pathway will now need to be discovered and/or characterized, including the identity of the Dpp8/9 substrate, how this substrate activates Nlrp1b to trigger pyroptosis, the role the proteasome plays in this pathway—an intriguing possibility is that the proteasome generates the Dpp8/9 substrate—and, perhaps most importantly, the PAMP that this pathway has been actually evolved to sense. On that note, Toxoplasma gondii, an obligate intracellular parasite (Ewald, et al., 2014; Gorfu, et al., 2014), was recently discovered to activate Nlrp1 to induce pyroptosis14,15. The molecular mechanism by which T. gondii activates Nlrp1 remains almost entirely unknown, but, like pyroptosis induced by Dpp8/9 inhibitors, it does not involve the N-terminal proteolysis of Nlrp1 (Cirelli, et al., 2014; Ewald, et al., 2014). Moreover, T. gondii, like Val-boroPro and unlike LT, activates pyroptosis in C57BL/6J macrophages, which contain allele 2 of Nlrp1b (Ewald, et al., 2014). As such, we speculate that it is possible that T. gondii and Dpp8/9 inhibitors might activate Nlrp1b through the same pathway, and perhaps Dpp8/9 even play a role in the detection of intracellular T. gondii infection. Regardless, our work here has identified a new cellular mechanism that activates Nlrp1, and we anticipate that these insights will inform future efforts to both understand the role of Nlrp1 in pathogen sensing and to modulate this pathway for therapeutic benefit.
SIGNIFICANCE
Val-boroPro, a small molecule inhibitor of post-proline cleaving serine proteases, induces immune-mediated tumor regressions in mouse models of cancer. Here we show that Val-boroPro stimulates the immune system by activating the Nlrp1b inflammasome. We also describe several key molecular features of this activation mechanism, including that 1) Nlrp1b autoproteolysis is required, 2) proteasome activity is required, 3) Val-boroPro activation of Nlrp1b does not involve direct proteolysis like anthrax lethal toxin, and 4) the expression of Nlrp1 and caspase-1 alone control cellular sensitivity to Val-boroPro. Overall, these data reveal an entirely new checkpoint controlling the activation of an important innate immune sensor.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Daniel Bachovchin (bachovcd@mskcc.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
HEK 293T (sex: female) and RAW 264.7 (sex: male) cells were purchased from ATCC and grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) at 37 °C in a 5% CO2 incubator. Cell lines were tested for mycoplasma using the MycoAlert™ Mycoplasma Detection Kit (Lonza).
Animal studies
11-week-old male C57BL/6J (The Jackson Laboratory), 129S6/SvevTac (Taconic), and Nlrp1b−/− (B6N.129S6-Nlrp1btm1Bhk/J, The Jackson Laboratory) mice were used in cytokine studies. This animal protocol was reviewed and approved by the Tufts University Medical Center Institutional Animal Care and Use Committee (IACUC). For comparison of cytokine induction in C57BL/6J and DBA/2J mice (both from The Jackson Laboratory), 7-week-old male animals were used. This animal protocol was reviewed and approved by the Memorial Sloan Kettering Cancer Center Institutional Animal Care and Use Committee (IACUC). Sample size was based on the statistical analysis of previous experiments with vehicle treated mice versus Val-boroPro treated mice (Adams, et al., 2004; Okondo, et al., 2017). No animals were excluded. Animals were randomly assigned to control and experimental groups. The investigators were not blinded.
METHOD DETAILS
Cloning
sgRNAs were designed using the Broad Institute’s web portal (Doench, et al., 2016) (http://www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design) and cloned into the lentiGuide-Puro vector (Addgene #52963) as described previously (Sanjana, et al., 2014). The sgRNA sequences used were: GFP_sg1 5′-GGGCGAGGAGCTGTTCACCG-3′; Nlrp1b_sg1 5′-TCCTGAGCTCTGTAATCACC-3′ (used in RAW 264.7 Nlrp1b KO1); Nlrp1b_sg2 5′-CCCCAATCACTAATGCCAGT-3′ (used in RAW 264.7 Nlrp1b KO2); Casp1_sg1 5′-TTAAACAGACAAGATCCTGA-3′ (used in RAW 264.7 Casp1 KO1), Nlrc4_sg1 5′-ACAGACGAGCCCTTATTCAA-3′ (used in RAW 264.7 Nlrc4 KO1 and 2). cDNA encoding the full length mouse Nlrp1b gene was cloned from RAW 264.7 macrophages and moved into the pLEX_307 vector (Addgene) and the pDEST27 vector (ThermoFisher Scientific) using Gateway technology (Thermo Fisher Scientific). The single amino acid point mutation S984A in Nlrp1b was generated with the QuickChange site-directed mutagenesis kit (Agilent). Cytosine 2178 within the PAM site of Nlrp1b sgRNA1 was also mutated to adenine in all Nlrp1b constructs to prevent their destruction in RAW 264.7 Nlrp1b KO1 cells. cDNA encoding mouse Casp1 were purchased from Origene. Casp1 was subcloned into a modified pLEX_307 vector with a hygromycin resistance marker.
Chemicals
Synthesis of the compounds VbP (Poplawski, et al.), compound 8j (Van Goethem, et al.), and 1G244 (Jiaang, et al.) were performed using the previously described synthetic methods. NMR spectra were recorded on a Bruker Advance 300 MHz NMR spectrometer employing a 5 mm inverse multinuclear probe, unless otherwise noted. Chemical shifts were reported in parts per million (δ) relative to DSS (in D2O) for 1H NMR. Mass spectra and HPLC retention times were recorded on a Hewlett Packard or Agilent 6110 LC/MSD system with UV detector (monitoring at 215 nm), using a Agilent Eclipse Plus C18 RP-HPLC column (4.6 × 50 mm, 1.8 μm) with solvent gradient A) water (0.1% TFA) and B) acetonitrile (0.08% TFA) at 0.5 ml/min. Unless otherwise noted, all HPLC retention times are given for an eluent gradient 2% B for the first 3 min, then from 2% to 98% B over 6 min, which was maintained for the next 5 min. The crude targets were purified by RP-HPLC using a Varian semi-preparative system with a Discovery C18 569226-U RP-HPLC column (25 cm × 21.2 mm, 5 μm) at 20 mL/min. All the purified compounds were analyzed by HPLC with purities >95%.
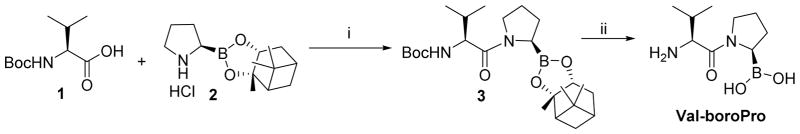
Synthesis of Val-boroPro
To a stirred solution of N-Boc-L-Valine 1 (2.2 g, 10 mmol) in anhydrous DMF (40 mL) was added N, N- diisopropylethylamine (DIEA, 3.8 mL, 21.3 mmol), HATU (4.0 g, 10.5 mmol) and L-boroPro-pn.HCl 2 (3.0 g, 10.5 mmol) sequentially at 0° under nitrogen. The cooling bath was removed and the resulting mixture was stirred at room temperature for 2 hr. The solvent was then removed in vacuo under 30°C. The residue was dissolved in ethyl acetate (200 mL), washed successively with KHSO4 (0.1 N, 3 × 30 mL), aq. NaHCO3 (5%, 3 × 30 mL), brine (2 × 20 mL) and dried with MgSO4, filtered. The solvent was removed in vacuo and the obtained crude product was purified by flash column chromatography over silica gel (3:1, hexanes/EtOAc) to afford compound 3 (4.1 g, 92%) as a white powder.
Compound 3 (2.2 g, 5 mmol) was dissolved in anhydrous dichloromethane (25.0 ml), cooled to −78 °C under Ar. BCl3 (1.0M in dichloromethane, 15 ml) was added dropwise and stirred for 1 hr at the same temperature. The reaction mixture was concentrated in vacuo, and co-evaporated to dryness with anhydrous methanol (2 × 20 ml). The residue was partitioned between water (20 ml) and ethyl ether (20 ml). The aqueous layer was separated, washed twice with more ethyl ether (2 × 20 ml), further purified by semi-preparative HPLC, and lyophilized to give the target product Val-boroPro (1.3 g as a TFA salt, 78%). 1 H NMR (D2O) δ 0.98 (d, J = 6.9 Hz, 3H), 1.08 (d, J = 6.9 Hz, 3H), 1.60 – 2.34 (m, 5H), 3.03 – 3.09 (m, 1H), 3.43 – 3.75 (m, 2H), 4.12 (d, J = 6.3 Hz, 1H). LC–MS (ESI+) m/z (rel intensity): 393.4 ([2 × (M - H2O) + H]+, 100); 197.2, ([M - H2O + H]+, 33). tR = 3.7 min.
For cell culture experiments, Val-boroPro was resuspended in DMSO containing 0.1% TFA to prevent compound cyclization.
Synthesis of 8j
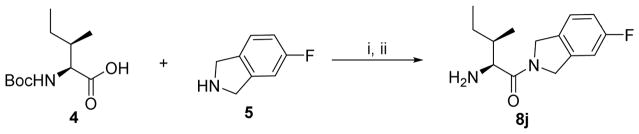
To a stirred solution of N-Boc-allo-Isoleucine 4 (0.7 g, 3 mmol) in anhydrous DMF (12 mL) was added N, N- diisopropylethylamine (DIEA, 1.2 mL, 6.4 mmol), HATU (1.2 g, 3.2 mmol) and 5-fluoroisoindoline.HCl 5 (0.54 g, 3.2 mmol) sequentially at 0° under nitrogen. The cooling bath was removed and the resulting mixture was stirred at room temperature for 2 hr. The solvent was then removed in vacuo under 30°C. The residue was dissolved in ethyl acetate (50 mL), washed successively with KHSO4 (0.1 N, 3 × 10 mL), aq. NaHCO3 (5%, 3 × 10 mL), brine (2 × 10 mL) and dried with MgSO4, filtered. The solvent was removed in vacuo and the obtained crude product was added to a stirred solution of 4N HCl in dioxane (7.5 mL, 10 eq.) while cooled at 0 to 5°C. After the addition, the reaction mixt ure is stirred for 3 hours at ambient temperature, and then concentrated in vacuo. The resulting solid was purified by semi-preparative HPLC and lyophilized to give the target product 8j (0.78 g as a HCl salt, 91%). 1 H NMR (D2O) δ 1.01 – 1.05 (m, 6H), 1.39 – 1.57 (m, 2H), 2.06 – 2.12 (m, 1H), 4.32 (d, J = 4.0 Hz, 1H), 4.64 – 5.01 (m, 4H), 7.04 – 7.10 (m, 2H), 7.27 – 7.34 (m, 1H). LC–MS (ESI+) m/z (rel intensity): 251.2 ([M + H] +, 100). tR = 8.6 min.
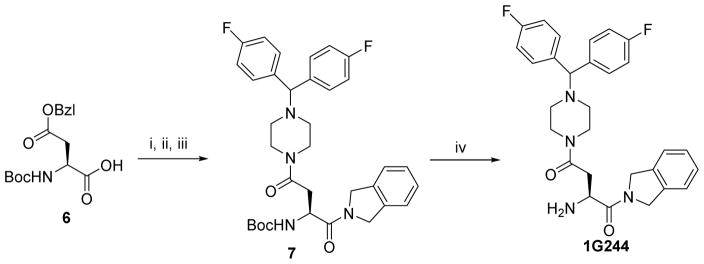
Synthesis of 1G244
To a stirred solution of N-Boc-L-Asp(OBzl)-OH 6 (1.6 g, 5 mmol) in anhydrous DMF (20 mL) was added N, N- diisopropylethylamine (DIEA, 1.9 mL, 10.6 mmol), HATU (2.0 g, 5.3 mmol) and isoindoline (0.63 g, 5.3 mmol) sequentially at 0° under nitrogen. The cooling bath was removed and the resulting mixture was stirred at room temperature for 2 hr. The solvent was then removed in vacuo under 30°C. The residue was dissolved in ethyl acetate (100 mL), washed successively with KHSO4 (0.1 N, 3 × 20 mL), aq. NaHCO3 (5%, 3 × 20 mL), brine (2 × 10 mL) and dried with MgSO4, filtered. The solvent was removed in vacuo and the obtained crude product was re-dissolved into MeOH (30 mL). 10% Pd-C (200 mg) was added and the resulting mixture was vigorously stirred under H2 (60 psi) overnight at room temperature. The mixture was then filtered through celite and washed with MeOH (2 × 10 mL). The combined solvent was removed in vacuo and the residual was re-dissolved into in anhydrous DMF (20 mL). N, N- diisopropylethylamine (DIEA, 1.9 mL, 10.6 mmol), HATU (2.0 g, 5.3 mmol) and 1-Bis(4-fluorophenyl)methyl piperazine (1.6 g, 5.5 mmol) sequentially at 0° under nitrogen. The cooling bath was removed and the resulting mixture was stirred at room temperature for 2 hr. The solvent was then removed in vacuo under 30°C. The residue was dissolved in ethyl acetate (100 mL), washed successively with KHSO4 (0.1 N, 3 × 20 mL), aq. NaHCO3 (5%, 3 × 20 mL), brine (2 × 10 mL) and dried with MgSO4, filtered. The solvent was removed in vacuo and the obtained crude product was purified by flash chromatography over silica gel to give Compound 7 (2.3 g, 75%). LC–MS (ESI+) m/z (rel intensity): 605.3 ([M + H] +, 100). tR = 9.8 min.
To a stirred ice-water cooled solution of 4N HCl in dioxane (5 mL, 20 eq.) was added Compound 7 (0.6 g, 1 mmol). After the addition, the reaction mixture is stirred for 3 hours at ambient temperature, and then concentrated in vacuo. The resulting solid was purified by semi-preparative HPLC and lyophilized to give the target product 1G244 (0.49 g as a HCl salt, 91%). 1H NMR (D2O) δ 3.03 – 3.25 (m, 2H), 3.25 – 3.35 (m, 4H), 3.75 – 3.85 (m, 4H), 4.70 – 5.08 (m, 5H), 5.44 (s, 2H), 7.15 – 7.20 (m, 4H), 7.35 – 7.40 (m, 4H), 7.61 – 7.65 (m, 4H). LC–MS (ESI+) m/z (rel intensity): 505.2 ([M + H] +, 100). tR = 8.8 min.
Knockout cell lines
Constructs were packaged into lentivirus in HEK 293T cells using the Fugene HD transfection reagent (Promega) and 2 μg of the vector, 2 μg psPAX2, and 0.2 μg pMD2.G, and incubated for 48 h. RAW 264.7 cells were spinfected with virus-containing supernatant from the transfection for 2 h at 1000g at 30 °C. After 2 days, cells were selected for stable expression of S. pyogenes Cas9 (Addgene #52962) using blasticidin (1 μg/mL) and for stable expression of sgRNAs using puromycin (5 μg/mL). After 10 d, single cells were isolated by serial dilution and expanded. To confirm allelic modifications, genomic DNA was harvested using Blood and Tissue Culture Kit (Qiagen). A 600 base pair region containing the sgRNA cut site was PCR amplified and subcloned using the CloneJET PCR Cloning Kit (ThermoFisher). Single bacterial colonies were isolated and sequenced.
LDH cytotoxicity assays
For experiments involving RAW 264.7 macrophages, cells were seeded at 0.5 × 106 cells/well in 6-well plates in standard growth medium 24 h prior to treatment. For ectopic expression of Nlrp1b in Nlrp1b−/− RAW264.7 cells, cells were nucleofected with 2 μg of a pLX-307 vector containing Nlrp1b WT or S984A, plated at 1 × 106 cells/well for 24 hours, and media was replaced prior to the indicated treatment. Cells were treated with the indicated concentrations of DMSO, Val-boroPro, 1G244, or 8j for 24 h, or treated with BSA or LT for 6 h unless indicated otherwise. Supernatants were then harvested and analyzed for LDH activity using an LDH cytotoxicity assay kit (Pierce). For experiments with proteasome inhibitors, RAW 264.7 macrophages were treated with bortezomib or MG132 30 minutes prior to the addition of Val-boroPro and/or LT, and then incubated for an additional 6 h. For LDH release experiments involving primary macrophages, mBMDM were isolated from the femurs and tibias of 7–9-week old mice, and 1 × 107 cells were plated on 10 cm petri dishes and differentiated in DMEM with 10% FBS and 20% L929 cell medium for 6 days. mBMDM were then scraped and reseeded at 0.5 × 106 cells/well in 12-well plates in Opti-MEM and incubated overnight before treatment. Cells were treated with the indicated concentrations of Val-boroPro or 8j for 24 h, or treated with BSA or LT for 6 h unless indicated otherwise. Supernatants were then harvested and analyzed for LDH activity as described above. This animal protocol was reviewed and approved by the Memorial Sloan Kettering Cancer Center Institutional Animal Care and Use Committee (IACUC). BALB/cJ, C57BL/6J, NOD/LtJ, DBA/2J, CAST/EiJ, Casp1−/− (B6N.129S2-Casp1tm1Flv/J), Nlrp1b−/− (B6N.129S6-Nlrp1btm1Bhk/J), and 129S1 (129S1/SvImJ) mice were purchased from The Jackson Laboratory. 129S6 (129S6/SvevTac) mice were purchased from Taconic. For LDH release experiments in HEK 293T cells, we first generated cells stably expressing mCasp1. HEK 293T cells were transfected 2 μg of the pLEX-307 containing mCasp1, 2 μg psPAX2, and 0.2 μg pMD2.G using Fugene HD transfection reagent (Promega) and incubated for 48 h. HEK 293T cells were then spinfected with virus-containing supernatant from the transfection for 2 h at 1000g at 30 °C supplemented 8 μg/mL polybrene. After 2 days, cells were selected for stable expression of mCasp1 using hygromycin (100 μg/mL). HEK 293T cells expressing mCasp1 were seeded at 0.5 × 106 cells/well in 6-well plates in standard growth medium, and transfected with 0.1 μg of the indicated Nlrp1b WT or Nlrp1b S984A vector and 1.9 μg of pLEX_307 containing GFP using the Fugene HD transfection reagent. After 24 h, cells were treated with Val-boroPro or LT as indicated and LDH release was assessed as described above.
Cytokine stimulation in mice
11-week-old male C57BL/6J (The Jackson Laboratory), 129S6/SvevTac (Taconic), and Nlrp1b−/− (B6N.129S6-Nlrp1btm1Bhk/J, The Jackson Laboratory) mice were fasted for 2 h prior to oral treatment with either vehicle (pH 2.0 water) or 100 μg Val-boroPro in pH 2.0 water (0.01N HCl). Serum was collected 6 h after dosing via cardiac puncture. G-CSF and CXCL1/KC levels were measured by Quantikine ELISA (R&D Systems). This animal protocol was reviewed and approved by the Tufts University Medical Center Institutional Animal Care and Use Committee (IACUC). For comparison of cytokine induction in C57BL/6J and DBA/2J mice (both from The Jackson Laboratory), 7-week-old animals were treated intraperitoneally with 100 μL of vehicle (1 mM HCl in PBS, pH = 7.4) or 100 μL of Val-boroPro (20 μg/100 μL). Val-boroPro was stored at 10x final concentration in 0.01N HCl and diluted into PBS immediately before dosing. Serum was collected 6 h after dosing via retro-orbital collection and G-CSF levels were measured by Quantikine ELISA (R&D Systems). This animal protocol was reviewed and approved by the Memorial Sloan Kettering Cancer Center Institutional Animal Care and Use Committee (IACUC). Sample size was based on the statistical analysis of previous experiments with vehicle treated mice versus Val-boroPro treated mice (Adams, et al., 2004; Okondo, et al., 2017). In these experiments, a sample size of 5 allows detection of increased G-CSF and CXCL1/KC levels with 80% power and a significance level below p = 0.05. No animals were excluded. The experiments were not randomized and the investigators were not blinded.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as mean values ± standard error of the mean (SEM). Levels of significance were determined by two-sided Student’s t-test for control versus experimental groups using GraphPad Prism (version 7). No analyses were performed to determine whether the data met the assumptions of this statistical approach. Statistical values, including the exact n and statistical significance, are reported in the Figure Legends.
Supplementary Material
Scheme 1.
Synthesis of Val-boroPro
Reagents and conditions: i. HATU, DIEA, DMF, 0°C to r.t, 92%; ii. BCl3 in CH2Cl2, −78°C, 78%.
Scheme 2.
Synthesis of 8j
Reagents and conditions: i. HATU, DIEA, DMF, 0°C to r.t; ii. 4N HCl in dioxane, 0°C to r.t, 91% for two steps.
Scheme 3.
Synthesis of 1G244
Reagents and conditions: i. Isoindoline, HATU, DIEA, DMF, 0°C to r.t; ii. H 2, 10% Pd-C, MeOH; iii. 1-Bis(4-fluorophenyl)methyl piperazine, HATU, DIEA, DMF, 0°C to r.t, 75% for 3 steps; iv. 4N HCl in dioxane, 0°C to r.t, 90%.
Okondo et al Highlights file.
Dpp8/9 inhibitors activate the Nlrp1b inflammasome to induce pyroptosis
Nlrp1b is polymorphic and sensitivity to Dpp8/9 inhibitors depends on the Nlrp1b allele
Dpp8/9 inhibitor-induced pyroptosis requires proteasome activity
Dpp8/9 inhibitors, unlike anthrax lethal toxin, do not activate Nlrp1b by proteolysis
Acknowledgments
We thank W. Bachovchin, W. Wu, and J. Lai for Val-boroPro, 1G244, and compound 8j. This work was supported by the Josie Robertson Foundation (D.A.B), a Stand Up to Cancer-Innovative Research Grant (Grant Number SU2C-AACR-IRG11-17 to D.A.B.; Stand Up to Cancer is a program of the Entertainment Industry Foundation. Research Grants are administered by the American Association for Cancer Research, the scientific partner of SU2C), the Pew Charitable Trusts (D.A.B. is a Pew-Stewart Scholar in Cancer Research), and the NIH (T32 GM115327-Tan to D.C.J. and the MSKCC Core Grant P30 CA008748).
Footnotes
AUTHOR CONTRIBUTIONS
M.O., S.D.R., C.Y.T., A.J.C., and D.C.J. performed experiments and analyzed data; S.E.P. performed the in vivo cytokine experiments with Nlrp1−/− mice; D.A.B. directed the project, analyzed data, and wrote the paper.
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams S, Miller GT, Jesson MI, Watanabe T, Jones B, Wallner BP. PT-100, a small molecule dipeptidyl peptidase inhibitor, has potent antitumor effects and augments antibody-mediated cytotoxicity via a novel immune mechanism. Cancer Res. 2004;64:5471–5480. doi: 10.1158/0008-5472.CAN-04-0447. [DOI] [PubMed] [Google Scholar]
- Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- Broz P, von Moltke J, Jones JW, Vance RE, Monack DM. Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe. 2010;8:471–483. doi: 10.1016/j.chom.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarria-Smith J, Mitchell PS, Ho AM, Daugherty MD, Vance RE. Functional and Evolutionary Analyses Identify Proteolysis as a General Mechanism for NLRP1 Inflammasome Activation. PLoS Pathog. 2016;12:e1006052. doi: 10.1371/journal.ppat.1006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarria-Smith J, Vance RE. Direct proteolytic cleavage of NLRP1B is necessary and sufficient for inflammasome activation by anthrax lethal factor. PLoS Pathog. 2013;9:e1003452. doi: 10.1371/journal.ppat.1003452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli KM, Gorfu G, Hassan MA, Printz M, Crown D, Leppla SH, Grigg ME, Saeij JP, Moayeri M. Inflammasome sensor NLRP1 controls rat macrophage susceptibility to Toxoplasma gondii. PLoS Pathog. 2014;10:e1003927. doi: 10.1371/journal.ppat.1003927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Osualdo A, Weichenberger CX, Wagner RN, Godzik A, Wooley J, Reed JC. CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PLoS One. 2011;6:e27396. doi: 10.1371/journal.pone.0027396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RC, Jin A, Rosales M, Yu S, Xia X, Ranola K, Schadt EE, Lusis AJ. A genome-wide set of congenic mouse strains derived from CAST/Ei on a C57BL/6 background. Genomics. 2007;90:306–313. doi: 10.1016/j.ygeno.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald SE, Chavarria-Smith J, Boothroyd JC. NLRP1 is an inflammasome sensor for Toxoplasma gondii. Infect Immun. 2014;82:460–468. doi: 10.1128/IAI.01170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger JN, Lich JD, Dare LC, Cook MN, Brown KK, Duraiswami C, Bertin J, Gough PJ. Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. J Biol Chem. 2012;287:25030–25037. doi: 10.1074/jbc.M112.378323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink SL, Bergsbaken T, Cookson BT. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc Natl Acad Sci U S A. 2008;105:4312–4317. doi: 10.1073/pnas.0707370105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frew BC, Joag VR, Mogridge J. Proteolytic processing of Nlrp1b is required for inflammasome activity. PLoS Pathog. 2012;8:e1002659. doi: 10.1371/journal.ppat.1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfu G, Cirelli KM, Melo MB, Mayer-Barber K, Crown D, Koller BH, Masters S, Sher A, Leppla SH, Moayeri M, et al. Dual role for inflammasome sensors NLRP1 and NLRP3 in murine resistance to Toxoplasma gondii. MBio. 2014:5. doi: 10.1128/mBio.01117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guey B, Bodnar M, Manie SN, Tardivel A, Petrilli V. Caspase-1 autoproteolysis is differentially required for NLRP1b and NLRP3 inflammasome function. Proc Natl Acad Sci U S A. 2014;111:17254–17259. doi: 10.1073/pnas.1415756111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmich KA, Levinsohn JL, Fattah R, Newman ZL, Maier N, Sastalla I, Liu S, Leppla SH, Moayeri M. Anthrax lethal factor cleaves mouse nlrp1b in both toxin-sensitive and toxin-resistant macrophages. PLoS One. 2012;7:e49741. doi: 10.1371/journal.pone.0049741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiaang WT, Chen YS, Hsu T, Wu SH, Chien CH, Chang CN, Chang SP, Lee SJ, Chen X. Novel isoindoline compounds for potent and selective inhibition of prolyl dipeptidase DPP8. Bioorg Med Chem Lett. 2005;15:687–691. doi: 10.1016/j.bmcl.2004.11.023. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- Kovarova M, Hesker PR, Jania L, Nguyen M, Snouwaert JN, Xiang Z, Lommatzsch SE, Huang MT, Ting JP, Koller BH. NLRP1-dependent pyroptosis leads to acute lung injury and morbidity in mice. J Immunol. 2012;189:2006–2016. doi: 10.4049/jimmunol.1201065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Levinsohn JL, Newman ZL, Hellmich KA, Fattah R, Getz MA, Liu S, Sastalla I, Leppla SH, Moayeri M. Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLoS Pathog. 2012;8:e1002638. doi: 10.1371/journal.ppat.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao KC, Mogridge J. Expression of Nlrp1b inflammasome components in human fibroblasts confers susceptibility to anthrax lethal toxin. Infect Immun. 2009;77:4455–4462. doi: 10.1128/IAI.00276-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman ZL, Printz MP, Liu S, Crown D, Breen L, Miller-Randolph S, Flodman P, Leppla SH, Moayeri M. Susceptibility to anthrax lethal toxin-induced rat death is controlled by a single chromosome 10 locus that includes rNlrp1. PLoS Pathog. 2010;6:e1000906. doi: 10.1371/journal.ppat.1000906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okondo MC, Johnson DC, Sridharan R, Go EB, Chui AJ, Wang MS, Poplawski SE, Wu W, Liu Y, Lai JH, et al. DPP8 and DPP9 inhibition induces pro-caspase-1-dependent monocyte and macrophage pyroptosis. Nat Chem Biol. 2017;13:46–53. doi: 10.1038/nchembio.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplawski SE, Lai JH, Li Y, Jin Z, Liu Y, Wu W, Wu Y, Zhou Y, Sudmeier JL, Sanford DG, et al. Identification of selective and potent inhibitors of fibroblast activation protein and prolyl oligopeptidase. J Med Chem. 2013;56:3467–3477. doi: 10.1021/jm400351a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyet JL, Srinivasula SM, Tnani M, Razmara M, Fernandes-Alnemri T, Alnemri ES. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001;276:28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- Squires RC, Muehlbauer SM, Brojatsch J. Proteasomes control caspase-1 activation in anthrax lethal toxin-mediated cell killing. J Biol Chem. 2007;282:34260–34267. doi: 10.1074/jbc.M705687200. [DOI] [PubMed] [Google Scholar]
- Taabazuing CY, Okondo MC, Bachovchin DA. Pyroptosis and Apoptosis Pathways Engage in Bidirectional Crosstalk in Monocytes and Macrophages. Cell Chem Biol. 2017;24:507–514. e504. doi: 10.1016/j.chembiol.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Leppla SH. Proteasome activity is required for anthrax lethal toxin to kill macrophages. Infect Immun. 1999;67:3055–3060. doi: 10.1128/iai.67.6.3055-3060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goethem S, Van der Veken P, Dubois V, Soroka A, Lambeir AM, Chen X, Haemers A, Scharpe S, De Meester I, Augustyns K. Inhibitors of dipeptidyl peptidase 8 and dipeptidyl peptidase 9. Part 2: isoindoline containing inhibitors. Bioorg Med Chem Lett. 2008;18:4159–4162. doi: 10.1016/j.bmcl.2008.05.079. [DOI] [PubMed] [Google Scholar]
- Van Opdenbosch N, Gurung P, Vande Walle L, Fossoul A, Kanneganti TD, Lamkanfi M. Activation of the NLRP1b inflammasome independently of ASC-mediated caspase-1 autoproteolysis and speck formation. Nat Commun. 2014;5:3209. doi: 10.1038/ncomms4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MP, Duncan B, Larabee S, Krauss A, Davis JP, Cui Y, Kim SY, Guimond M, Bachovchin W, Fry TJ. Val-boroPro accelerates T cell priming via modulation of dendritic cell trafficking resulting in complete regression of established murine tumors. PLoS One. 2013;8:e58860. doi: 10.1371/journal.pone.0058860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong FL, Mamai O, Sborgi L, Boussofara L, Hopkins R, Robinson K, Szeverenyi I, Takeichi T, Balaji R, Lau A, et al. Germline NLRP1 Mutations Cause Skin Inflammatory and Cancer Susceptibility Syndromes via Inflammasome Activation. Cell. 2016;167:187–202. e117. doi: 10.1016/j.cell.2016.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.