Abstract
A recent study by Waller and colleagues evaluated the reliability, specificity, and generalizability of using functional connectivity data to identify individuals from a group. The authors note they were able to replicate identification rates in a larger version of the original Human Connectome Project (HCP) dataset. However, they also report lower identification accuracies when using historical neuroimaging acquisitions with low spatial and temporal resolution. The authors suggest that their results indicate connectomes derived from historical imaging data may be similar across individuals, to the extent that this connectome-based approach may be inappropriate for precision psychiatry and the goal of drawing inferences based on subject-level data. Here we note that the authors did not take into account factors affecting data quality and hence identification rates, independent of whether a low spatiotemporal resolution acquisition or a high spatiotemporal resolution acquisition is used. Specifically, we show here that the amount of data collected per subject and in-scanner motion are the predominant factors influencing identification rates, not the spatiotemporal resolution of the acquisition. To do this, we investigated identification rates in the HCP dataset as a function of the amount of data and motion. Using a dataset from the Consortium for Reliability and Reproducibility (CoRR), we investigated the impact of multiband versus non-multiband imaging parameters; that is, high spatiotemporal resolution versus low spatiotemporal resolution acquisitions. We show scan length and motion affect identification, whereas the imaging protocol does not affect these rates. Our results suggest that motion and amount of data per subject are the primary factors impacting individual connectivity profiles, but that within these constraints, individual differences in the connectome are readily observable.
Introduction
A key goal of precision psychiatry is leveraging individual differences in neuroimaging data to generate predictive models related to behavior. As highlighted by Waller et al. (2017), finding reliable markers across datasets remains an important part of this process. As such, they investigate the generalizability of a previous method using functional connectivity fMRI data to identify individuals from a group (‘connectome fingerprinting’; Finn et al., 2015). Waller et al. demonstrate that identification can be replicated in the same high spatiotemporal resolution dataset (i.e. acquired using multiband acquisition sequences), consistent with other work to replicate the method (Finn et al., 2017; Kaufmann et al., 2017; Vanderwal et al., 2017), though they note lower accuracies using a dataset acquired with lower spatiotemporal resolution (i.e. acquired using non-multiband acquisition sequences). In addition, the authors also show that the specificity of the identification procedure is lower when a within-subject correlation threshold is introduced into the ID pipeline. Therefore, the authors argue that the identification method may not generalize to datasets with lower spatiotemporal resolution because individual features may only be detectable in data acquired with high spatiotemporal resolution. However, in their study, the authors did not take into account other factors affecting data quality and hence the identification process, namely scan duration and subject motion. Here we evaluate the impact of not only spatiotemporal resolution during image acquisition, but also other data quality factors on identification rates.
Methods
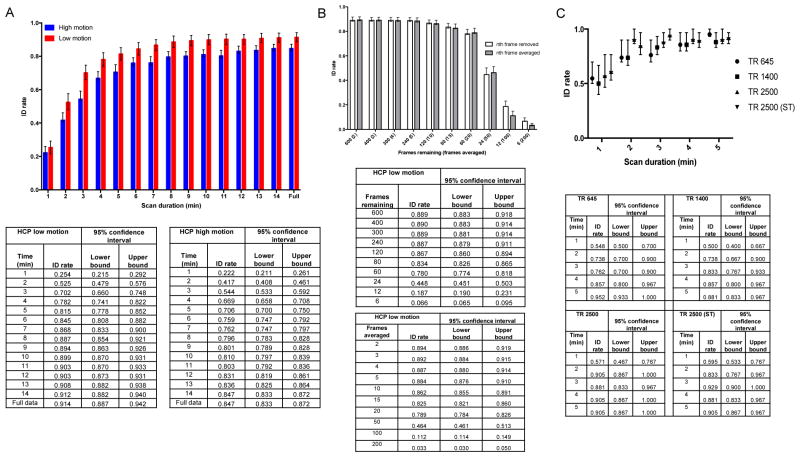
The HCP 900 subjects release (Van Essen et al., 2013) was used to investigate scan time and motion. Data were pre-processed and connectivity matrices were calculated as described elsewhere (Finn et al., 2015; Finn et al., 2017; Shen et al., 2017). All analyses were performed using the left-to-right (LR) phase encoding rest runs from days one and two. Of note, HCP TR = 720 ms. To study motion, subjects were separated into low and high motion groups using a mean frame-frame displacement threshold of 0.1mm averaged over the two sessions. Of the 819 subjects available with all data and day one and two LR rest scans, 603 were in the low motion group and 216 subjects were in the high motion group. To study the effect of scan time, we truncated time courses to correspond to the number of frames in 1, 2,…,14 minutes and calculated connectivity matrices from the shortened data. Because of the difference in sample sizes among the low and high motion groups, we repeatedly subsampled 216 subjects in the low motion group and performed identification 1000 times. The mean ID rate and 95% confidence intervals were therefore calculated from the subsampled data (Fig 1A). To investigate when ID rates plateaued, we used the Levenberg-Marquardt nonlinear least squares algorithm to fit the following nonlinear regression model function: , where t = time, IDrate = ID rate at time t, maxIDrate = maximum ID rate determined by the model, and x = time required for the ID rate to reach approximately 63% of its maximum value. We defined plateauing of the ID rate to be the time points when the rate was 95 and 99% of the maximum ID rate.
Figure 1.
The effect of scan duration, motion, and differences in spatiotemporal resolution on identification rates. (A) Top: Separating the HCP 900 subjects into groups based on motion and performing identification with increasing amounts of data. Identification rate for each group is indicated at each scan duration time. Note that the high and low motion groups have the same scan durations at a given time point on the x-axis. Both groups have equal sample sizes (n = 216). Red and blue bars represent the low and high motion groups, respectively. (B) Top: Simulating the effects of a lower TR in the HCP. Data from all 603 low motion subjects were subsampled (white bar); x-axis indicates the number of frames remaining. In a separate analysis every n adjacent frames were averaged; (grey bars); x-axis indicates in parentheses the number of adjacent frames used to average. (C) Top: Identification rates achieved using multiband or non-multiband imaging parameters to assess the effect of spatiotemporal resolution. Multiband imaging was performed on groups labelled as TR 645 and TR 1400; TR 2500 was acquired via non-multiband imaging; TR 2500 (ST) indicates these subjects underwent slice-time correction. Identification rate achieved for each scanning protocol is indicated at each scan duration time. Error bars correspond to 95% confidence intervals. Note that in (A), (B), and (C), the lower part of each panel includes the actual ID rate obtained and the 95% confidence intervals.
In a separate analysis (Fig 1B), we subsampled data (after low-pass filtering; approximate cutoff frequency of 0.12 Hz) from each of the 603 low motions subjects to simulate the effects of lower sampling frequencies (longer TR) versus total amount of scan time. For this analysis, we selected n frames from the duration of a subject’s time course such that sampling every other frame produced 600 frames of the original 1200; sampling every 3rd frame resulted in 400 frames remaining, etc. Hence, these subsampled data still spanned the same overall acquisition time window. It should also be noted that this subsampled data has lower signal to noise ratio (SNR) than real data acquired at a longer TR because of the additional T1 recovery that would occur with a longer TR. Connectivity matrices were subsequently calculated from the subsampled data. In addition, we performed a follow-up analysis using a similar strategy except that instead of removing every nth frame we averaged data from every n adjacent frames to again simulate a slower sampling frequency (Fig 1B) and boost the SNR.
To study the effect of spatiotemporal resolution (Fig 1C), we utilized a publically available test-retest dataset from the Nathan Kline Institute (NKI; http://fcon_1000.projects.nitrc.org/indi/CoRR/html/nki_1.html). This dataset contains individuals scanned with both multiband and non-multiband acquisition sequences, thus allowing us to investigate the impact of different pulse sequences on ID rates. Data acquisition parameters have been described previously (Liao et al., 2013). Briefly, three resting-state fMRI sequences were obtained for each of the 24 subjects: 1) multiband scan with TR=645 ms; 2) multiband scan with TR=1400 ms; and 3) and a non-multiband echo planar imaging (EPI) scan with TR=2500 ms. One subject was excluded due to brain atrophy (subject 0021001); one subject was excluded due to excessive head motion (3795193; greater than 3 degrees rotation); and we were unable to locate session 2 data for subject 6471972, leaving 21 subjects in the final analysis. We did not apply a further motion cutoff with these subjects due to the small sample size. The preprocessing steps have been previously described (Noble et al., 2017), except we performed skull-stripping using optiBET (Lutkenhoff et al., 2014). Though we did not perform slice-time correction on the multi-band data, we performed analyses on the TR=2500 subjects with and without slice-time correction.
The identification procedure was carried out as described previously using Matlab code released by Finn et al. (2015) and utilized by Waller and colleagues (https://www.nitrc.org/frs/?group_id=51). To directly compare our results to the main findings of Waller et al. we used the same subset of nodes from the frontoparietal and medial frontal networks to perform identification. To generate 95% confidence intervals, we calculated bootstrapped identification accuracies by subsampling approximately 70% of the subjects in each condition tested.
Results
Using only the edges derived from the frontoparietal and medial frontal networks, we found identification was affected by both motion and total scan time in the HCP 900 dataset. Given the differences in sample sizes of the datasets used in this study and that of the authors (BLP 85), ID rates should not be directly compared to those obtained by Waller et al. Rather our focus is on the relative importance of motion, scan time, and acquisition sequence.
We observed that identification rates were consistently higher for subjects in the low motion group compared to the high motion group (P < 0.05; Fig 1A) with the only exception being when one minute of data was used. Increasing scan duration, or the amount of data per subject, resulted in increasing rates of identification for both groups. Using nonlinear regression, we determined that ID rates reached 95% and 99% percent of maximum at approximately 6.85 and 10.3 minutes, respectively, for the low motion group, and at 8.7 and 14.4 minutes, respectively, for the high motion group, supporting the notion that increasing scan durations past 3.7 and 5.2 minutes (the scan times of 2/3 scans used in Waller et al.) results in higher ID rates for both groups, and that for high motion subjects, more data is needed to achieve a successful identification.
To further investigate the importance of total scan time, we subsampled the HCP 900 data from the 603 low motion subjects, in one case by taking every other volume, and in the other case by averaging adjacent frames, to simulate a lower TR. We found identification rates were relatively stable after both procedures (Fig 1B) and did not begin to decrease until removing every 20th frame (60 frames remaining; P < 0.05) and averaging over every 50 frames (P < 0.05).
We next used the NKI dataset to evaluate the effect of spatiotemporal resolution on identification rates. Using the same framework as above, we found that identification rates were effectively the same across imaging conditions at a given time point (P > 0.05; Fig 1C). Similar to the HCP results above, we observed that increasing the scan duration increased identification rates in the NKI dataset. Interestingly, it was scan duration, and not number of samples, that had the largest effect on identification rates. In other words, a shorter TR cannot compensate for a shorter acquisition time: in general, it is better to have fewer samples distributed across a longer temporal window than more samples acquired in quick succession (echoing results in Laumann et al., 2015, Airan et al., 2016 and Noble et al., 2017).
General comments
The study by Waller et al. (2017) provides a valuable contribution to the goal of precision psychiatry by replicating the original connectome fingerprinting work, and we applaud their efforts to expand the method to other datasets and generalize the findings to larger groups of subjects. However, the authors point out that accuracy and specificity are expected to drop with lower quality datasets (in the authors’ words, datasets of “standard quality,” as opposed to “high quality” datasets like the HCP). While we agree that identification rates are expected to drop as sample size increases, higher identification accuracies can still be obtained by improving factors other than the particular EPI acquisition used. Here, we have demonstrated that scan duration and subject movement both affect identification rates, but were unable to find an effect of the particular EPI acquisition strategy. Given the known effect of motion on estimates of functional connectivity (Power et al., 2015; Satterthwaite et al., 2012), it is reasonable that high motion subjects might be harder to identify from scan to scan. Further, the findings with respect to EPI protocol (i.e. spatiotemporal resolution) are generally consistent with the work of Airan et al. (2016), in which they applied a non-parametric measure to study the differentiation of subjects in the NKI dataset and observed no clear relationship regarding multiband versus non-multiband data. Taking all of these factors into account, we suggest that if Waller et al. (2017) had applied a similar motion threshold to the BLP 85 dataset and if they had longer scan durations per subject, they would have higher identification rates—even with their standard quality fMRI data.
In a more general sense, our results reinforce the importance of the amount of data collected per subject in detecting between-subject differences. In the original connectome fingerprinting article it was shown that identification rates increase with increasing data (Finn et al., 2015); follow-up work similarly demonstrated identification is affected by time (Finn et al., 2017). Other studies have also shown that longer acquisition times and more data are associated with increases in the test-retest reliability of functional connectivity measures (Birn et al., 2013; Mueller et al., 2015; Shah et al., 2016; Noble et al., 2017; Laumann et al., 2015) as well as estimates of individual differentiation (Airan et al., 2016).
We note that the authors showed that the specificity of the ID method could be low even when high accuracies are obtained. While the specificity findings are noteworthy and we appreciate their work, it is not clear to what extent specificity in the context of identification-based studies is necessary. Identifiability of the functional connectome by itself is interesting, yet identification provides no information related to behavioral/cognitive measures, disease course, etc. The goal is the development of connectome-based predictive models that the unique patterns of connectivity provide (Shen et al., 2017). Identification per se is not the primary objective, and thus identifying a patient with high confidence from scan to scan does not inform patient response to treatment or disease prognosis.
With this in mind, we reiterate an important aspect of our original work: capitalizing on individual variability in functional connectomes and using this to build meaningful models predicting some cognitive feature as opposed to only identification of participants. Prediction of individual features is a valuable objective for neuroimaging (Gabrieli et al., 2015) and our previous work (Finn et al., 2015; Rosenberg et al., 2017) supports the notion that it is possible to generate predictive models of behavior from neuroimaging data. While we agree with Waller et al. that further developments are needed to yield clinically available biomarkers based on individual connectomes, there are several emerging promising results suggesting the potential clinical utility (Drysdale et al., 2017) of this approach. In this development, attention to the impact of factors such as motion, scan duration, and acquisition parameters is a key part of the process. We maintain that establishing the link between individual connectomes and behavior is an important goal for precision psychiatry. We thank Waller et al. for extending the work of our identification method, and we look forward to further developments in using individual connectome data in precision psychiatry.
Acknowledgments
This work was supported by a Medical Scientist Training Program training grant (NIH/NIGMS T32GM007205; C.H.) and the US National Science Foundation Graduate Research Fellowship under grant number DGE1122492 (S.M.N.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Airan RD, Vogelstein JT, Pillai JJ, Caffo B, Pekar JJ, Sair HI. Factors affecting characterization and localization of interindividual differences in functional connectivity using MRI. Hum Brain Mapp. 2016;37:1986–1997. doi: 10.1002/hbm.23150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM, Molloy EK, Patriat R, Parker T, Meier TB, Kirk GR, Nair VA, Meyerand ME, Prabhakaran V. The effect of scan length on the reliability of resting-state fMRI connectivity estimates. Neuroimage. 2013;83:550–558. doi: 10.1016/j.neuroimage.2013.05.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, Fetcho RN, Zebley B, Oathes DJ, Etkin A, Schatzberg AF, Sudheimer K, Keller J, Mayberg HS, Gunning FM, Alexopoulos GS, Fox MD, Pascual-Leone A, Voss HU, Casey BJ, Dubin MJ, Liston C. Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med. 2017;23:28–38. doi: 10.1038/nm.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn ES, Scheinost D, Finn DM, Shen X, Papademetris X, Constable RT. Can brain state be manipulated to emphasize individual differences in functional connectivity? Neuroimage. 2017 doi: 10.1016/j.neuroimage.2017.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, Constable RT. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat Neurosci. 2015;18:1664–1671. doi: 10.1038/nn.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrieli JD, Ghosh SS, Whitfield-Gabrieli S. Prediction as a humanitarian and pragmatic contribution from human cognitive neuroscience. Neuron. 2015;85:11–26. doi: 10.1016/j.neuron.2014.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann T, Alnaes D, Doan NT, Brandt CL, Andreassen OA, Westlye LT. Delayed stabilization and individualization in connectome development are related to psychiatric disorders. Nat Neurosci. 2017;20:513–515. doi: 10.1038/nn.4511. [DOI] [PubMed] [Google Scholar]
- Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen MY, Gilmore AW, McDermott KB, Nelson SM, Dosenbach NU, Schlaggar BL, Mumford JA, Poldrack RA, Petersen SE. Functional System and Areal Organization of a Highly Sampled Individual Human Brain. Neuron. 2015;87:657–670. doi: 10.1016/j.neuron.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao XH, Xia MR, Xu T, Dai ZJ, Cao XY, Niu HJ, Zuo XN, Zang YF, He Y. Functional brain hubs and their test-retest reliability: a multiband resting-state functional MRI study. Neuroimage. 2013;83:969–982. doi: 10.1016/j.neuroimage.2013.07.058. [DOI] [PubMed] [Google Scholar]
- Lutkenhoff ES, Rosenberg M, Chiang J, Zhang K, Pickard JD, Owen AM, Monti MM. Optimized brain extraction for pathological brains (optiBET) PLoS One. 2014:e115551. doi: 10.1371/journal.pone.0115551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Wang D, Fox MD, Pan R, Lu J, Li K, Sun W, Buckner RL, Liu H. Reliability correction for functional connectivity: Theory and implementation. Hum Brain Mapp. 2015;36:4664–4680. doi: 10.1002/hbm.22947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S, Spann MN, Tokoglu F, Shen X, Constable RT, Scheinost D. Influences on the Test-Retest Reliability of Functional Connectivity MRI and its Relationship with Behavioral Utility. Cereb Cortex. 2017:1–15. doi: 10.1093/cercor/bhx230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Schlaggar BL, Petersen SE. Recent progress and outstanding issues in motion correction in resting state fMRI. Neuroimage. 2015;105:536–551. doi: 10.1016/j.neuroimage.2014.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MD, Finn ES, Scheinost D, Constable RT, Chun MM. Characterizing Attention with Predictive Network Models. Trends Cogn Sci. 2017;21:290–302. doi: 10.1016/j.tics.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Wolf DH, Loughead J, Ruparel K, Elliott MA, Hakonarson H, Gur RC, Gur RE. Impact of in-scanner head motion on multiple measures of functional connectivity: relevance for studies of neurodevelopment in youth. Neuroimage. 2012;60:623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah LM, Cramer JA, Ferguson MA, Birn RM, Anderson JS. Reliability and reproducibility of individual differences in functional connectivity acquired during task and resting state. Brain Behav. 2016;6:e00456. doi: 10.1002/brb3.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Finn ES, Scheinost D, Rosenberg MD, Chun MM, Papademetris X, Constable RT. Using connectome-based predictive modeling to predict individual behavior from brain connectivity. Nat Protoc. 2017;12:506–518. doi: 10.1038/nprot.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K Consortium WUMH. The WU-Minn Human Connectome Project: an overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderwal T, Eilbott J, Finn ES, Craddock RC, Turnbull A, Castellanos FX. Individual differences in functional connectivity during naturalistic viewing conditions. Neuroimage. 2017;157:521–530. doi: 10.1016/j.neuroimage.2017.06.027. [DOI] [PubMed] [Google Scholar]
- Waller L, Walter H, Kruschwitz JD, Reuter L, Muller S, Erk S, Veer IM. Evaluating the replicability, specificity, and generalizability of connectome fingerprints. Neuroimage. 2017;158:371–377. doi: 10.1016/j.neuroimage.2017.07.016. [DOI] [PubMed] [Google Scholar]