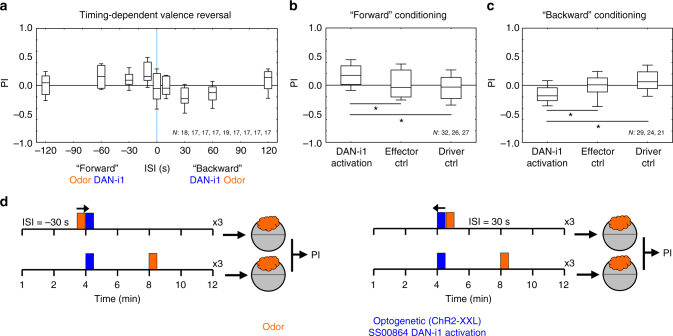
Fig. 6.
Timing-dependent valence reversal of optogenetic DAN-i1 reinforcement. a Larvae of the experimental genotype (heterozygous for SS00864-Gal4 and UAS-ChR2-XXL) were trained by presenting an odor and optogenetically activating DAN-i1, at various relative timings. A 30-s odor presentation either preceded a 30-s DAN-i1 activation (forward conditioning, plotted as negative inter-stimulus interval, ISI) or the odor followed DAN-i1 activation (backward conditioning, positive ISI). Subsequently, larvae were tested for their odor preference. For short-interval forward conditioning, positive scores reveal appetitive memory. For short-interval backward conditioning, negative scores indicate aversive memory. b Appetitive memory scores were confirmed after forward conditioning (ISI of −10 s) for the experimental genotype (heterozygous for SS00864-Gal4 and UAS-ChR2-XXL (DAN-i1 activation)). Control larvae heterozygous for only UAS-ChR2-XXL (effector Ctrl) or SS00864-Gal4 (driver Ctrl) did not show any memory. c Aversive memory scores were also confirmed after backward conditioning (ISI of 30 s) for the experimental genotype (DAN-i1 activation). Again, genetic controls did not show any memory. The preference scores underlying the associative performance indices can be found in Supplementary Fig. 11. Sample sizes are indicated within the figure. Other details as in Fig. 3. d Schematic of the timed odor-DAN-i1 association protocol. Orange and blue colors indicate the timing of odor presentation and optogenetic activation of DAN-i1, respectively, during each of the three 12-min training trials. In the example timeline, paired training (top) involves intervals between the onset of odor presentation and optogenetic activation of −30 s (left) and 30 s (right); the timing of events for unpaired training is shown to the bottom. After either paired or unpaired training, larvae are tested for their preference for the odor; the associative performance indices (PIs) are then calculated from the difference in preference between paired-trained and unpaired-trained larvae