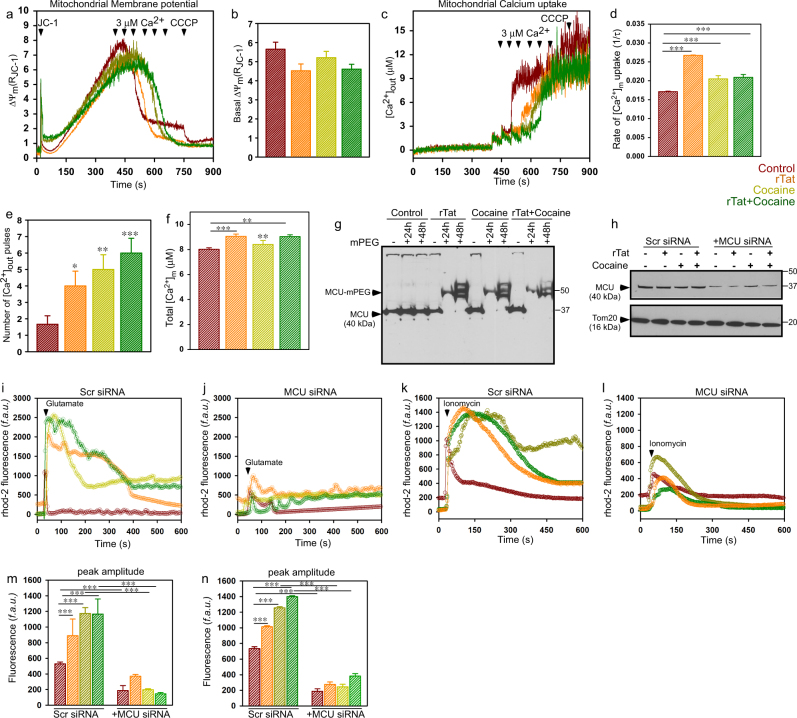
Fig. 2. MCU-mediated Ca2+ uptake enhances mitochondrial metabolism in astrocytes exposed to rTat/cocaine.
a, c Representative Δψm (a) and [Ca2+]out traces (c). Permeabilized cells were loaded with the Δψm indicator JC-1 and extra-mitochondrial Ca2+ ([Ca2+]out) indicator Fura2FF to which a series of extra-mitochondrial Ca2+ pulses (3 μM) were added (arrowheads) to assess the [Ca2+]out clearance rate. b Quantification of basal Δψm before the addition of [Ca2+]out. d–f Quantification of the rate of [Ca2+]m uptake as a function of decrease in [Ca2+]out (d), number of extra-mitochondrial Ca2+ pulses handled (e), Total [Ca2+]m taken up by astrocytes treated with rTat, cocaine, or rTat and cocaine (f). g rTat/cocaine-induced oxidative modification of MCU. Representative Western blot of lysates prepared from astrocytes exposed to rTat, cocaine, or rTat and cocaine and expressing MCU-FLAG (AdMCU). The lysates were incubated with mPEG5 (30 mins) and oxidation of MCU was identified using anti-Flag antibody. h Cell lysates prepared from control (Scr siRNA) and MCU KD astrocytes exposed to rTat, cocaine, or rTat and cocaine were assessed by Western blotting with anti-MCU antibody. The outer mitochondrial membrane receptor, TOM20 was used as the loading control. i, j Mean traces of [Ca2+]m (rhod-2) dynamics in control (Scr siRNA) (i) and MCU KD (j) astrocytes exposed to rTat, cocaine, or rTat and cocaine. After measurement of baseline fluorescence, cells were stimulated with glutamate (200 μM, arrowheads) and changes in [Ca2+]m fluorescence were measured. k, l Mean traces of [Ca2+]m (rhod-2) dynamics in control (Scr siRNA) (k) and MCU KD (l) astrocytes exposed to rTat, cocaine, or rTat and cocaine. After measurement of baseline fluorescence, cells were stimulated with Ionomycin (2.5 μM, arrowheads) and changes in [Ca2+]m fluorescence were measured. m Quantification of peak amplitude of rhod-2 fluorescence after glutamate stimulation. n Quantification of peak amplitude of rhod-2 fluorescence after ionomycin stimulation. Data represents Mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001; n = 6-8. Scr siRNA is a Scrambled/non-target siRNA control.