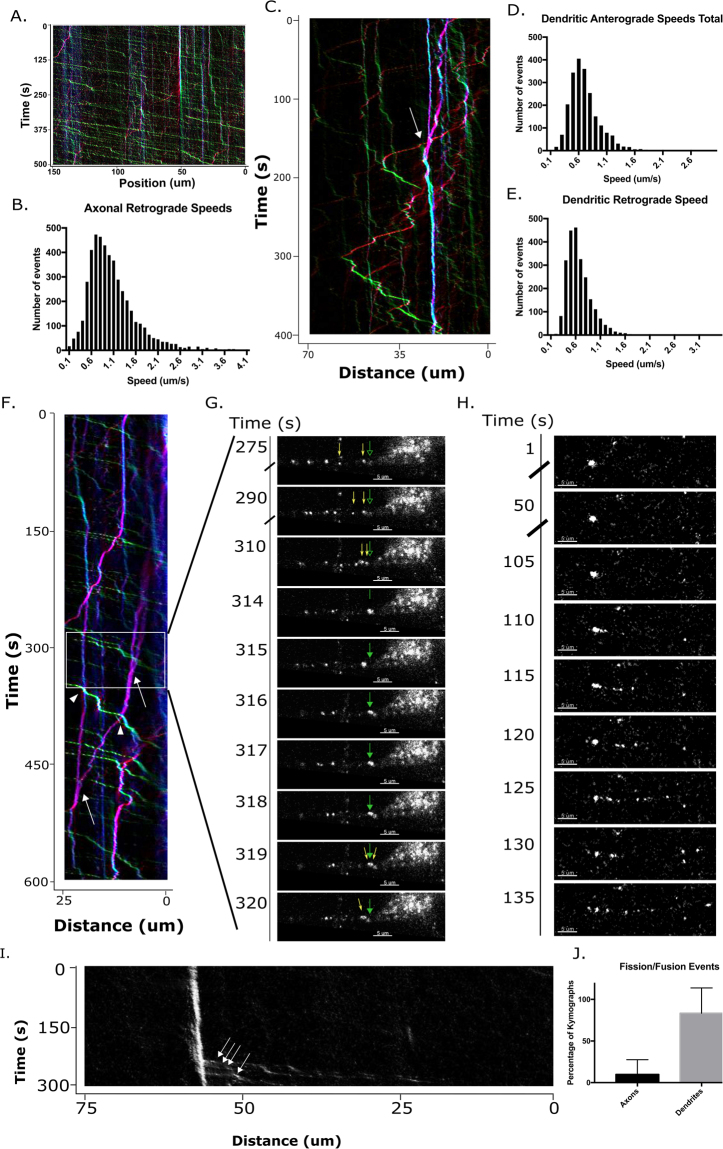
Figure 3.
SE movement in dendrites. (A) Representative kymograph of retrograde axonal movement of signaling endosomes (visualized with Cy3-anti-FLAG antibody fed in distal chamber). Images were taken at a rate of 1 frame/sec. Kymographs were generated by KymoClear in which stationary events are coded in blue, retrograde events in green, and anterograde events in red. (B) Axonal retrograde speeds were calculated from 34 of movies from 3 independent experiments and show an average speed of 0.99 µm/s. (C) Representative kymograph of SE movements in a dendrite. Arrow indicates multiple endosomes coming together in a putative fusion event. (D) Histogram of anterograde dendritic speeds. Speeds were calculated from 3 independent experiments and show an average speed of 0.71 ± 0.28 µm/s. (E) Histogram of retrograde dendritic speeds. Speeds were calculated from 3 independent experiments and show an average speed of 0.67 ± 0.26 µm/s. (F) Representative kymograph showing multiple putative fusion events in a single dendrite. Arrows indicate putative fusion between SEs, arrowheads point to an SE that passes with no fusion. Box indicates position of still images in (G). (G) Stills from dendritic movie showing putative fusion event. Green arrows point at position of putative fusion, and yellow arrows indicate individual endosomes dynamically merging and splitting. (H,I) A stationary SE undergoing multiple fission events is shown as stills from a dendritic movie (H) and as a kymograph (I). (J) Quantification of putative fission/fusion events in axons vs dendrites. n = 15 cells for axons; 13 cells for dendrites; 3 independent experiments each.