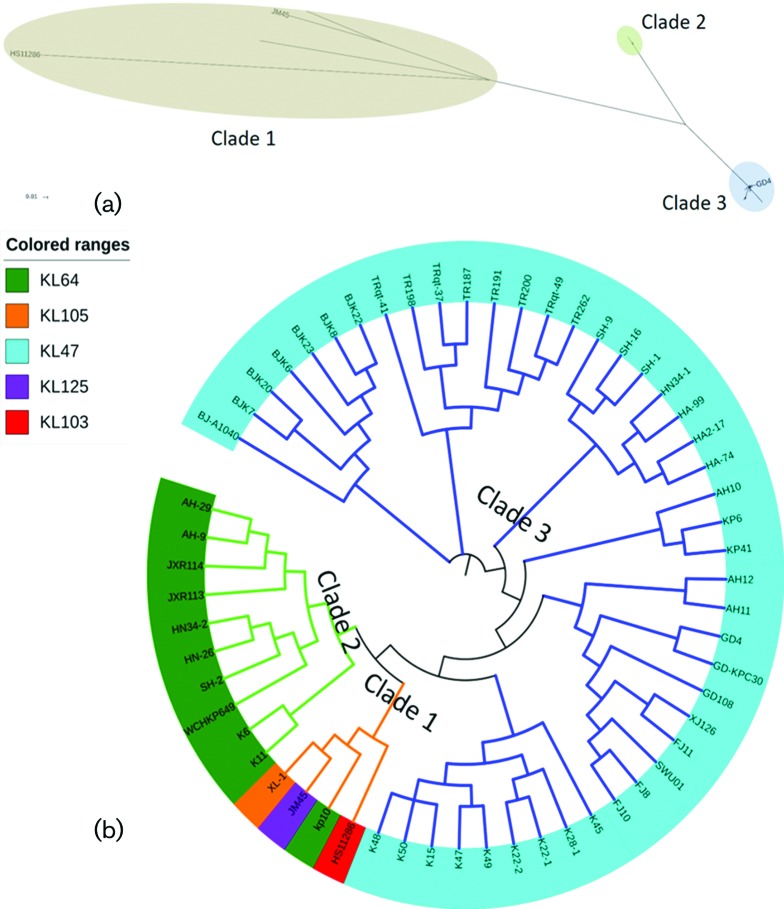
Fig. 2.
Approximately maximum-likelihood phylogeny estimated with the parsnp software [28] based on a total of 6749 unique concatenated SNPs in the core genome of 58 clinical ST11 K. pneumoniae strains from China. (a) Unrooted phylogenetic analysis of 58 K. pneumoniae clinical isolates. GD4, HS11286 and JM45 are ST11 strains with completed genome sequences. Bar, 0.01 substitutions per nucleotide position. (b) Circular phylogenetic tree of the 58 strains. The lengths of the branches are not proportional to the evolutionary distances in the cladogram. Different background colors underneath each strain name indicate the K locus type of the corresponding strain detected by Kaptive [32].