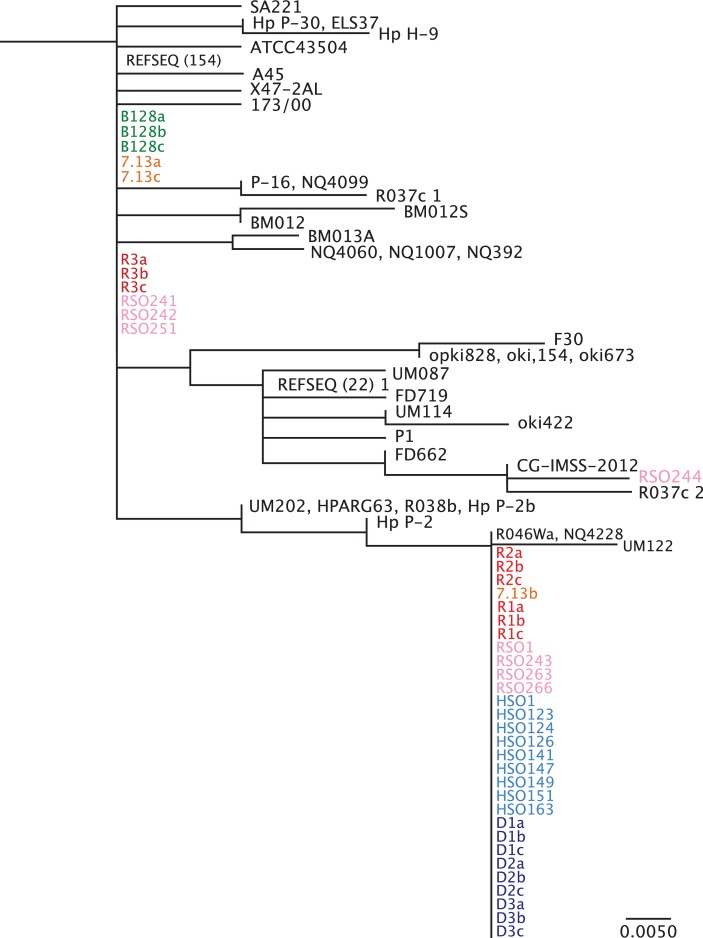
Figure 3.
Phylogenetic analysis of Fur amino acid sequence. Phylogenetic analysis was performed for full-length Fur from all available Helicobacter pylori sequence data within the National Center for Biotechnology Information NCBI database. H. pylori single colonies from strains B128 (green) and 7.13 (orange), and in vivo-adapted strains isolated from gerbils maintained on iron-replete (red) and iron-depleted (blue) diets as well as in vivo-adapted strains isolated from gerbils maintained on regular salt (pink) and high salt (light blue) diets were included in these analyses. R1-3 and D1-3 designate independent gerbils maintained on either iron-replete (n=3) or iron-depleted (n=3), respectively, while a–c designate different single colonies isolated from each gerbil. HSO and RSO designate independent high salt output and regular salt output H. pylori strains, respectively. REFSEQ (#) represents the number of unique H. pylori sequences from different H. pylori strains that cluster under the same phylogeny. Although all Fur sequences from the NCBI database were used, this phylogenetic analysis shows only the branch to which the newly sequenced strains cluster. The scale bar represents amino acid changes per site.