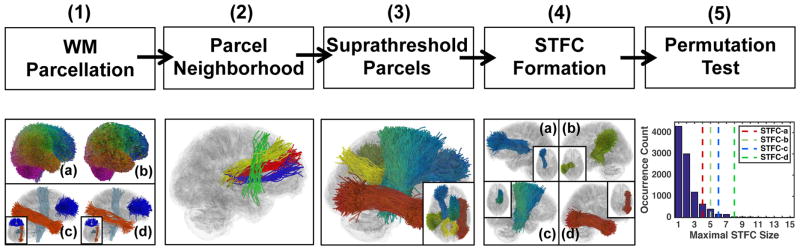
Figure 1.
Method overview: (1) WM Parcellation: (a) and (b) show the study-specific data-driven groupwise WM parcellation (atlas) and a subject-specific WM parcellation; (c) and (d) show several example atlas WM tract parcels and the corresponding subject-specific WM tract parcels (d); (2) Parcel Neighborhood: example neighbors of the red tract parcel include the yellow and the blue parcels but not the more distant green one with dissimilar WM anatomy to the others; (3) Suprathreshold Parcels: example fiber parcels that passed a parcel-level statistic test from the whole brain according to a quantitative parameter extracted from each parcel (the diffusion feature of return-to-the-origin probability (RTOP) was used in this study), computed using a Student’s t-test given a user-given threshold (uncorrected p-value = 0.05 is used in this study); (4) STFC Formation: example STFCs obtained using clique percolation (CP) to join neighboring parcels with uncorrected significance; (5) Permutation Test: example null distribution obtained from a non-parametric permutation test with summary statistic of maximal STFC size. For each STFC, its corrected significance is computed by comparing its STFC size (dashed lines) to the null distribution.