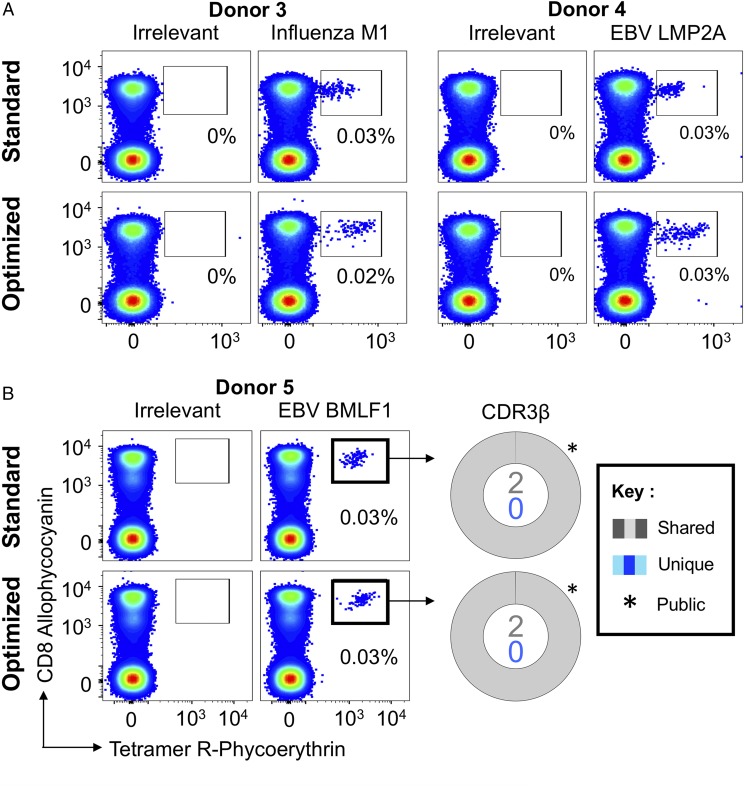
FIGURE 2.
Ex vivo detection of Ag-specific CD8 T cells using standard pMHC multimer staining technology. (A) PBMCs from HLA A2+ healthy donors were stained with tetramers bearing the influenza M158–66 peptide (GILGFVFTL; left panel) or the EBV LMP2A426–434 peptide (CLGGLLTMV; right panel), using tetramer alone (standard) or in combination with PKI and anti-fluorochrome Ab (optimized). Gates were set on single lymphocytes and live CD3+CD14negCD19neg cells. Irrelevant tetramer made with preproinsulin-derived peptide15–24 (ALWGPDPAAA) (65) or human telomerase reverse transcriptase (hTERT540–548, ILAKFLHWL) was used to set the gates. The percentage of CD8+Tet+ T cells is shown for each gate. (B) HLA A2+ PBMCs were sorted using EBV BMLF1280–288 (GLCTLVAML) standard tetramer staining and optimized staining protocols (left panel). CD8+Tet+ T cells were sorted, and TCR β-chain sequencing was performed to compare clonotype capture between protocols. TCR β-chains (right panel) are displayed as sort-shared (gray) or sort-unique (blue) sections of a pie, with each section for each sort corresponding to a different CDR3β. The number of shared (gray) and unique (blue) CDR3s for the respective sorts are shown in the center of each pie. TRBV usage is shown in Supplemental Fig. 2. A complete list of sorted β clonotypes can be found at https://github.com/antigenomics/vdjdb-db/issues/243.