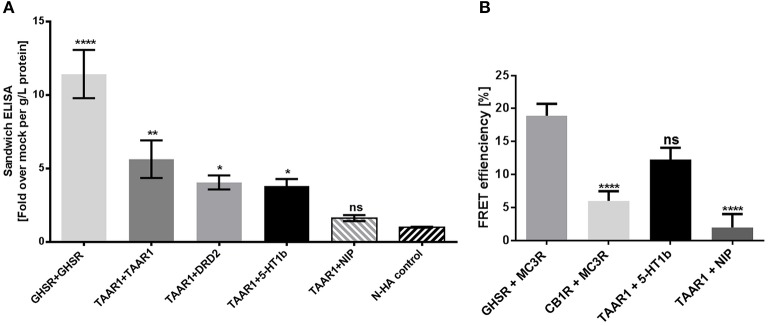
Figure 4.
TAAR1 and 5-HT1b can interact. Sandwich ELISA and FRET were performed to investigate dimerization. (A) For sandwich ELISA, receptor pairs were overexpressed in COS-7 cells, receptors were NHA- and FLAG-tagged and vice versa. The homodimers GHSR and TAAR1, as the heterodimer TAAR1 and DRD2 were used as positive control; TAAR1 and rM3R were used as negative control. NHA control are cells over expressing NHA-tagged TAAR1, 5-HT1b, DRD2, and GHSR. Data are shown as mean ± SEM of fold over mock transfection. Data were pooled from four to eight independent assays measured in three replicates. For statistical analysis, a one-way ANOVA was performed in relation to mock transfection. (B) To determine FRET efficiency, HEK293 overexpressed receptor pairs, one being CFP and one the YFP tagged and vice versa. The heterodimer GHSR and MC3R was used as positive control, CB1R and MC3R as negative control. rM3R was used as NIP for TAAR1. Compared to GHSR and MC3R dimers, the negative control and TAAR1 with the NIP showed no protein-protein-interactions, whereas TAAR1 and 5-HT1b co-expression did. Data were pooled from 11 to 19 cells and a one-way ANOVA was performed and the mean of each column was compared with the mean of all other columns. Statistical significance was defined as *p ≤ 0.05, **p ≤ 0.01, ****p ≤ 0.0001.