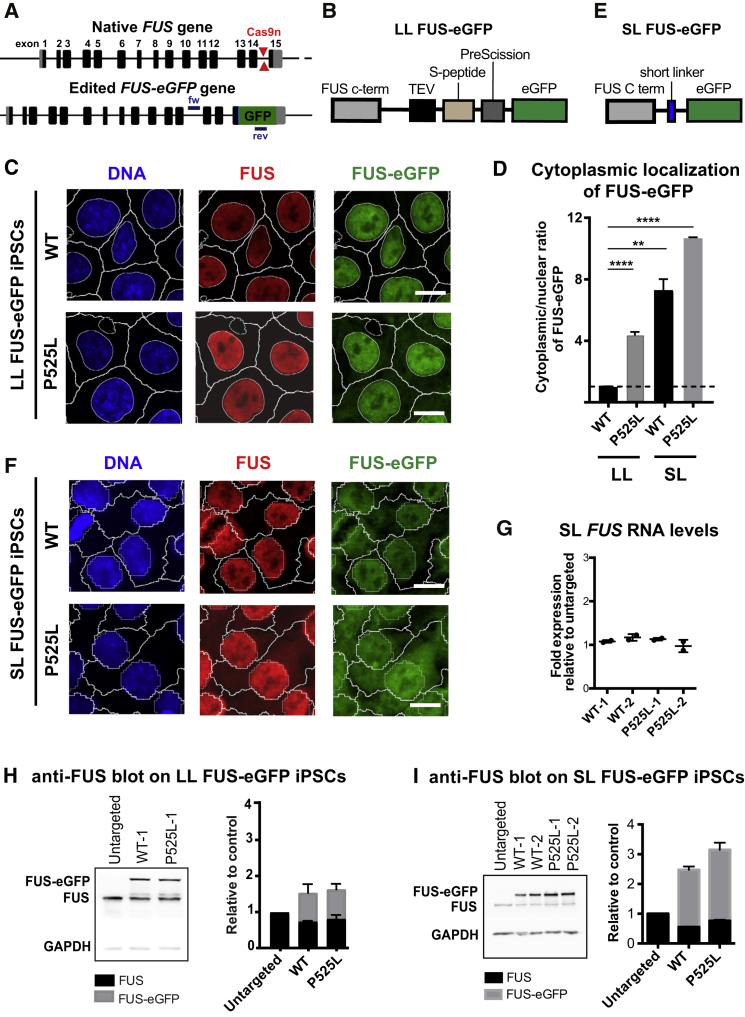
Figure 1.
Generation of FUS-eGFP Reporter iPSCs
(A–F) Examples of FUS gene before and after gene editing to insert eGFP (A). Cas9n cut sites are shown with locations of primers used for genotyping. Schematic representation of the linkers used to knock in eGFP in LL (B) and SL (E) iPSC lines. Confocal micrographs showing FUS localization in WT and P525L FUS-eGFP cells for LL (C) and SL (F). Nuclear and cytoplasmic segmentation is displayed. Scale bar, 10 μm. (D) Quantification of the FUS-eGFP cytoplasmic-nuclear ratio in WT and P525L FUS-eGFP LL and SL iPSCs. All values are expressed as fold relative to LL WT FUS-eGFP iPSCs. Data represent results from three independent measurements. Error bars indicate SD. ∗∗p < 0.01 and ∗∗∗∗p < 0.0001.
(G–I) qRT-PCR showing total FUS RNA levels (G). WB and relative quantification using an anti-FUS antibody showing expression of FUS and FUS-eGFP in LL (H) and SL (I) iPSCs. Results are shown for two different WT and P525L lines as biological replicates and were reproduced in three independent experiments. Error bars indicate SD.