Figure 2.
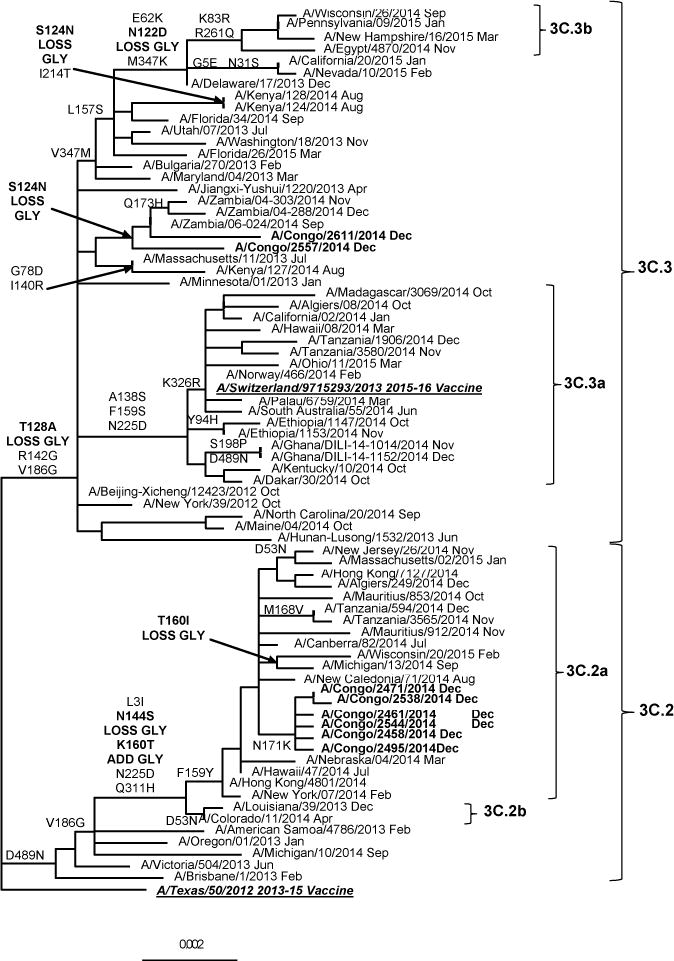
Representative hemagglutinin ML phylogenetic tree of A(H3N2) viruses collected since 2013. The A/Texas/50/2012 (underlined, bold) was used as reference in TREESUB to estimate ML phylogenetic trees via RAxML and PAML, and for automated annotation of amino acid substitutions in the nexus format. The consensus HA tree and the transcribed amino acid substitutions were visualized in FigTree. The scale bar represents the average number of nucleotide substitutions per site. Genetic groups 3C.2 and 3C.3, with the respective subgroups are represented in the tree. DRCongo isolates (bold) grouped with 3C.2a and 3C.3 reference viruses. Other African isolates clustered with the A/Switzerland/9715293/2013 (underlined, bold) vaccine virus from genetic group 3C.3a.
