Abstract
Cr(VI)-containing compounds are well-established lung carcinogens. Chronic exposure of the normal human epithelial cells is able to induce malignant cell transformation, the first stage of metal carcinogenesis. These Cr(VI)-transformed cells exhibit increased level of antioxidants, reduced capacity of generating reactive oxygen species (ROS), and development of apoptosis resistance, promoting tumorigenesis of Cr(VI)-transformed cells, the second stage of metal carcinogenesis. The mechanism of Cr(VI) induced carcinogenesis is still under investigation. Recent studies indicate that ROS play a positive role in the first stage while a negative role in the second stage. Transformed cells adapt metabolism to support tumor initiation and progression. Altered metabolic activities directly participate in the process of cell transformation or support a large requirement for nucleotides, amino acids, and lipids for tumor growth. In malignantly Cr(VI)-transformed cells, mitochondrial oxidative phosphorylation is defective, and pentose phosphate pathway, glycolysis, and glutaminolysis are upregulated. These metabolic reprogramming supports rapid cell proliferation and contributes to tumorigenesis of Cr(VI)-transformed cells. This article summarizes the current progress in the studies of metabolic reprogramming and Cr(VI) carcinogenesis with emphasis on the metabolic enzymes and oxidative stress related major oncogenic pathways.
Keywords: Oxidative stress, Chromium (VI), Carcinogenesis, metabolic reprogramming
Introduction
When normal cells are converted to malignant transformed cells and progress to cancer, the metabolism is altered. In contrast to the normal differentiated cells which rely mainly on mitochondrial oxidative phosphorylation (OXPHOS) for generation of needed energy, cancer cells depend on anaerobic glycolysis, a phenomenon called “the Warburg effect” for energy. This altered cellular metabolism, also called metabolic reprogramming, is recognized as one of the cancer phenotypes. Accumulative evidence reveals that various oncogenic pathways are involved in the metabolic regulation. Expression of glucose transporters and glycolytic enzymes are increased in numerous cancers and may contribute to tumor progression [1, 2]. It has been reported that oncogenes and tumor suppressor genes, such as hypoxia inducible factor 1 (HIF-1) [3], c-Myc [4–6], p53 [7.8], and PI3K/Akt [9], directly promote metabolism of glucose and glutamine.
Chromate (Cr(VI)) compounds, widely used in industry, have been shown to be toxic and carcinogenic on humans [10–13]. Cr(VI) is structurally similar to sulfate and phosphate anions; therefore, it readily enters into the cells via non-specific anion transporters [14]. Once inside the cells, Cr(VI) undergoes a series of metabolic reductions and forms intermediate Cr species, including Cr(V) and Cr(IV), and is finally reduced to Cr(III) [14, 15]. In the Cr(VI) reduction process, reactive oxygen species (ROS) are produced, resulting in oxidative DNA damage. The intermediates Cr(V), Cr(IV), and the final product Cr(III) are very reactive, causing Cr-DNA adducts and genomic alterations. Epidemiological studies have shown that occupational exposure to Cr(VI) is associated with a high rate of lung cancer in workers employed in these industries [10–13]. Environmental Cr(VI) exposure is also a public health concern and is associated with long-term carcinogenic effects of the lung [12, 16]. The mechanisms of Cr(VI) carcinogenesis have not yet been fully understood, it generally believed that ROS are important in inducing malignant cell transformation, the first stage of metal carcinogenesis [3–5, 17*]. ROS can be involved in various carcinogenic processes [17*]. Recent studies from our laboratories have shown that once cells are malignantly transformed, the capacity of those transformed cells to generate ROS is sharply reduced, contributing to the development of apoptosis resistant and subsequent tumorigenesis [18, 19*]. Thus, the decreased ROS generation in Cr(VI)-transformed cells is oncogenic in promoting tumorigenesis, the second stage of metal carcinogenesis. The oncogenic role of ROS in the first stage of Cr(VI) carcinogenesis (malignant cell transformation) and anti-oncogenic role in the second stage (tumorigenesis) reflects the metabolic reprogramming during the change from normal cells to malignantly transformed cells. Although this reprogramming may play an important role in the mechanism of metal carcinogenesis in general and Cr(VI) carcinogenesis in particular, its underlying mechanism remains to be investigated. This article provides an outline on progress and future perspectives in oxidative stress and metabolic reprogramming in Cr(VI) carcinogenesis.
Glycolysis
Glucose homeostasis is controlled by glycolysis/OXPHOS and gluconeogenesis pathway. Glycolysis is the enzymatic conversion of glucose into lactate, which produces 2 ATP per glucose molecule. In the presence of oxygen, normal cells primarily adopt mitochondrial OXPHOS to produce 36 ATP per glucose molecule. Cancer cells favor aerobic glycolysis over OXPHOS to meet their energy demand, suggesting that cancer cells are adapted to survive and proliferate in the absence of mitochondrial ATP production. Mitochondria play a major role as energy suppliers and ROS regulators. Although various mechanisms of carcinogenesis induced by Cr(VI) have been demonstrated, it is generally believed that Cr(VI)-induced oxidative stress is important in converting normal cells to malignantly transformed cells. It has been reported that Cr(VI) suppressed all five mitochondrial complexes involved in OXPHOS in a variety of model systems with higher potency of complexes I, II, and V than complexes III and IV. Our recent study has observed that in Cr(VI)-transformed cells mitochondrial ATP production was reduced and non-mitochondrial oxygen consumption was increased, indicating the defect of mitochondrial ATP production [20**]. The results from RNA sequencing analysis show that levels of various enzymes involved in all five complexes were reduced in Cr(VI)-transformed cells compared to passage-matched normal cells (Table 2), demonstrating that Cr(VI)-transformed cells are defective in mitochondrial ATP production.
Table 2.
Relative level of enzymes involved in oxidative phosphorylation in Cr(VI)-transformed cells and passage-matched normal cells
| Gene Symbol | Mitochondrial complex | BEAS-2B | BEAS-2B-Cr(VI) |
|---|---|---|---|
| NDUFA13 | Complex I | 58.1 ± 6.2 | 34.9 ± 12.2 |
| NDUFA11 | Complex I | 10.0 ± 0.3 | 6.4 ± 1.0 |
| NDUFC2 | Complex I | 154.6 ± 10.8 | 81.3 ± 23.5 |
| NDUFS8 | Complex I | 182.5 ± 2.5 | 99.2 ± 26.7 |
| NDUFS7 | Complex I | 71.7 ± 3.8 | 46.4 ± 13.4 |
| NDUFS5 | Complex I | 544.2 ± 77.2 | 271.8 ± 21.4 |
| NDUFB7 | Complex I | 254.3 ± 20.1 | 153.6 ± 29.6 |
| NDUFV1 | Complex I | 12.4 ± 4.0 | 5.8 ± 1.2 |
| NDUFA6 | Complex I | 105.5 ± 3.4 | 0.5 ± 0.8 |
| NDUFA2 | Complex I | 152.4 ± 26.5 | 72.6 ± 20.1 |
| SDHA | Complex II | 246.5 ± 45.9 | 105.6 ± 20.1 |
| UQCRC1 | Complex III | 108.1 ± 8.3 | 84.5 ± 18.0 |
| UQCR11 | Complex III | 160.3 ± 18.5 | 101.5 ± 4.1 |
| UQCR10 | Complex III | 103.6 ± 11.4 | 61.4 ± 9.1 |
| UQCRH | Complex III | 42.0 ± 3.8 | 26.0 ± 2.1 |
| COX4I1 | Complex IV | 11.6 ± 4.6 | 3.4 ± 1.3 |
| COX6A1 | Complex IV | 1.4 ± 0.6 | 0.7 ± 0.1 |
| COX6B1 | Complex IV | 6.4 ± 1.7 | 1.8 ± 0.7 |
| ATP5H | Complex V | 631.8 ± 39.9 | 362.8 ± 11.1 |
| ATP5L | Complex V | 119.5 ± 11.7 | 87.3 ± 19.9 |
| ATP5C1 | Complex V | 76.2 ± 19.2 | 1.6 ± 0.6 |
Relative levels of enzymes involved in mitochondrial oxidative phosphorylation. The method used is the same as that in Table 1. Genes involved in mitochondrial oxidative phosphorylation were detected using EBseq. A false detection rate analysis with 0.05 threshold was performed and considered as biostatistic difference (p<0.05). Data represent mean ± SD (n=3).
Aerobic glycolysis, maximizing ATP production, does not require an increase in mitochondrial capacity [21]. Cr(VI)-transformed cells generated more lactate without significant changes in glucose uptake and ATP production, indicating a switch from mitochondrial respiration to glycolysis [20**]. The results from Table 1 show that several glycolysis enzymes including ADP-specific glucokinase (ADPGK), enolase 1 (ENO1), hexokinase (HK2), phosphoglycerate kinase (PGK1), dihydrolipoamide S-acetyltransferase (DLAT), pyruvate dehydrogenase E1 (PDHA1), glucose-6-phosphatase 3 (G6PC3), pyruvate kinase (PKM), aldolase A (ALDOA), and phosphofructokinase (PFKM) were upregulated in Cr(VI)-transformed cells, indicating that Cr(VI)-transformed cells utilize glycolysis for energy production under defective mitochondrial function. It should be noticed that many metabolic enzymes are regulated through allostery and/or post-translation, such as pyruvate dehydrogenase, a complex with multiple subunits and cofactors, whose activity is regulated by phosphorylation/dephosphorylation. Thus, characterization of metabolic flux together with transcriptomics is more appropriate to evaluate the metabolic changes upon Cr(VI) exposure. Cancer stem cells (CSCs) or cancer-initiating cells, a small subset of malignant cells that exhibit high capacity of self-renewal and differentiation, have been reported to utilize aerobic glycolysis for biosynthesis and energy requirement [22]. About 1% of Cr(VI)-transformed cells have been identified as CSCs. These CSCs are metabolic inactive as evidenced by dramatic reductions of glucose uptake, lactate production, and ATP content [20**]. These small population of CSCs may be the driving force for the increased glycolysis of Cr(VI)-transformed cells.
Table 1.
Relative level of metabolic enzymes in Cr(VI)-transformed cells and passage-matched normal cells
| Gene Symbol | Target pathway | BEAS-2B | BEAS-2B-Cr(VI) |
|---|---|---|---|
| ADPGK | Glycolysis | 0.4 ± 0.6 | 6.7 ± 3.2 |
| ALDOA | PPP/Glycolysis | 6.8 ± 5.3 | 254.9 ± 57.0 |
| DLAT | Glycolysis | 14.7 ± 1.3 | 21.6 ± 2.6 |
| EN01 | Glycolysis | 1.5 ± 0.8 | 71.6 ± 0.9 |
| G6PC3 | Glycolysis | 50.7 ± 4.1 | 97.3 ± 14.9 |
| GPI | Glycolysis | 19.1 ± 4.8 | 73.8 ± 8.9 |
| HK2 | Glycolysis | 21.2 ± 4.7 | 32.1 ± 2.8 |
| PDHA1 | Glycolysis | 48.9 ± 5.5 | 158.5 ± 11.7 |
| PFKM | Glycolysis | 29.5 ± 5.8 | 83.9 ± 3.6 |
| PGK1 | Glycolysis | 107.5 ± 44.6 | 221.7 ± 68.5 |
| PKM | Glycolysis | 112.5 ± 86.5 | 261 ± 73.9 |
| G6PD | PPP | 29.2 ± 6.5 | 142.3 ± 50.2 |
| PGD | PPP | 6.8 ± 3.7 | 31.4 ± 5.9 |
| PRPS1 | PPP | 59.4 ± 2.3 | 135.2 ± 8.1 |
| PRPS2 | PPP | 27.2 ± 0.1 | 53.6 ± 5.9 |
| RPE | PPP | 28.1 ± 2.1 | 39.0 ± 4.7 |
| RPIA | PPP | 151.9 ± 39.7 | 324.3 ± 85.5 |
| TALD01 | PPP | 28.8 ± 8.1 | 55.8 ± 7.7 |
| TKT | PPP | 151.9 ± 39.7 | 324.3 ± 85.5 |
| GFPT1 | Glutaminolysis | 14.4 ± 1.7 | 26.8 ± 6.7 |
| GLS | Glutaminolysis | 19.3 ± 4.6 | 30.3 ± 10.4 |
| GOT2 | Glutaminolvsis | 99.4 ± 32.8 | 283.9 ± 36.7 |
Relative levels of metabolic enzymes involved in glycolysis, PPP, and glutaminolysis. Normal BEAS-2B cells (BEAS-2B) and Cr(VI)-transformed cells (BEAS-2B–Cr) were cultured in 10-cm dishes. After 90% of confluence, the cells were subjected for extraction and purification of RNA using RNAeasy mini kit. Whole transcriptome sequencing analysis was performed using HiSeq 2500 Rapid Run. Differentially expressed genes involved in glycolysis, PPP, and glutaminolysis were detected using EBseq. A false detection rate analysis with 0.05 threshold was performed and considered as biostatistic difference (p<0.05). Data represent mean ± SD (n=3).
Energy metabolism is a balanced mechanism controlled by catabolic (glycolysis and oxidative phosphorylation) and anabolic (gluconeogenesis) reactions. Fructose-1, 6-bisphosphatase (FBP1), a rate limiting enzyme in gluconeogenesis, catalyzes the hydrolysis of fructose-1, 6-bisphosphate to fructose 6-phosphate and inorganic phosphate. It has been reported that loss of FBP1 is correlated with advanced stage and poor prognosis of cancer [23, 24]. Inhibition of FBP1 increased glucose uptake and lactate secretion in HK-2 human renal cells and in consistent, forced expression of FBP1 reduced glucose uptake, lactate secretion, and glucose-derived TCA cycle intermediates in renal carcinoma RCC10 cells [25*]. In CSCs low level of FBP1 is beneficial for cancer cell growth due to (a) inductions of superiority of glycolysis and increased glucose uptake, facilitating the production of glycolysis intermediates and the energy supply during hypoxia and (b) inhibition of ROS generation induced by mitochondrial complex 1, protecting cells from oxidative stress [26]. In Cr(VI)-transformed cells FBP1 level is low compared to that in passage-matched normal cells and FBP1 is lost in CSCs [20**]. Ectopic expression of FBP1 in CSCs reduced glucose uptake, lactate production, and glycolysis [20**], indicating that FBP1 plays an important role in glucose metabolism.
HIF-1α is important in angiogenesis and in cancer development [27–29]. Its level is elevated in more than half of human cancers and their metastases [30–35]. The occurrence of Warburg effect indicates the activation of oncogenic signaling, such as hypoxia inducible factor (HIF)-1α, resulting in promotion of glucose uptake and anabolic metabolism [21]. This transcription factor upregulates many glycolytic enzymes, in which their gene promoters contain consensus binding motif 5’-(C/G/T)ACGTGC(G/T)-3’ of HIF-1α. HIF-1α protein is rapidly degraded at normoxia via pVHL-mediated ubiquitin-proteasome pathway, whereas hypoxia blocks degradation of HIF-1α protein, leading to its accumulation [36]. Stabilization of HIF-1α modulates metabolic adaptation to low molecular oxygen levels through increase of cellular glycolysis [37]. HIF-1α upregulates glucose transporters and glycolytic enzymes [38*]. HIF1α also upregulates pyruvate dehydrogenase kinases (PDKs), the enzymes control entry of glucose-derived pyruvate into tricarboxylic acid (TCA) cycle [39, 40]. HIF-1α was activated in Cr(VI)-repeatedly exposed cells [41] or in Cr(VI)-transformed cells [42]. HIF-1α is able to bind to five glycolytic enzymes including phosphofructokinase (PFK), aldolase (ALDA), phosphoglycerate kinase 1 (PGK1), enolase 1 (ENO1), pyruvate kinase (PKM), and lactate dehydrogenase (LDHA) [43]. The results from Table 1 show that these five enzymes are upregulated in Cr(VI)-transformed cells. It is very likely that HIF-1α directly binds to the promoters of these glycolytic enzymes. FBP1 binds to HIF-1α inhibitory domain, blocking its induction of glycolysis [25*]. Thus, reduced FBP1 level in Cr(VI)-transformed cells may induce glycolysis through decreased binding to HIF1α.
Phosphorylation on metabolic enzymes also contributes to aerobic glycolysis [44]. PI3K/Akt phosphorylates hexokinase and PFK-2 [45] and promotes GLUT expression and plasma membrane localization [46], suggesting that activation of PI3K/Akt pathway promotes the Warburg effect by stimulating glucose uptake and further catabolism by glycolysis. It has been demonstrated that PI3K/Akt/p38 MAPK is responsible for HIF-1α activation in Cr(VI)-transformed cells [41], indicating involvement of PI3K/Akt/p38 in the upregulation of glycolysis of Cr(VI)-transformed cells.
Pentose phosphate pathway (PPP)
Pentose phosphate pathway (PPP), a classic metabolic pathway, consists of oxidative and non-oxidative branches. The oxidative PPP converts glucose-6-phosphate (G6P), a glycolytic intermediate, into ribulose-5-phosphate and generates NADPH, which is used for glutathione production, detoxification, and biosynthesis of lipids. The non-oxidative branch involves reversible carbon-exchanging reactions with the final products as fructose-6-phosphate and glyceraldehyde-3-phosphate, which participate in glycolysis and downstream metabolic pathways [47*]. PPP is upregulated in many types of tumors [47*, 48]. The activities of glucose-6-phosphate dehydrogenase (G6PD) and transketolase (TKT), key PPP enzymes, were increased in cancer cells [49, 50]. An early study indicated that short-term exposure of human erythrocytes to Cr(VI) did not exhibit effect on any of the three PPP enzymes, glucose-6-phosphate dehydrogenase (G6PDH), 6-phosphogluconate dehydrogenase (PGD), and transsketolase (TKT) [51]. We have performed transcriptomics by RNA sequencing analysis. The results show that in Cr(VI)-transformed cells expressions of several PPP enzymes including phosphorybosyl pyrophosphate synthase 1 & 2 (PRPS1/2), G6PD, ribulose 5-phosphate 3-epimerase (RPE), transaldolase (TALDO1), PGD, ribose 5-phosphate isomerase (RPIA), aldolase A (ALDOA), and TKT were elevated compared to passage-matched normal cells (Table 1), indicating that PPP is upregulated in Cr(VI)-transformed cells.
Nuclear factor (erythroid-derived 2)-like 2 (Nrf2) is a key transcription factor that regulates antioxidant proteins to neutralize ROS and to restore cellular redox balance [52*, 53]. This transcript factor plays dual roles in carcinogenesis. In normal cells, activation of inducible Nrf2 decreases malignant cell transformation via decrease of oxidative stress [54**–56]. Conversely, in cancer cells constitutively activated Nrf2 exerts oncogenic effects by protecting these cells from oxidative stress and chemotherapeutics [57*, 58]. Constitutive activation of Nrf2 has been identified in several types of human cancer cell lines and tumors [54**, 59–61]. Cancers with high Nrf2 level are associated with poor prognosis [62, 63], resistance to therapeutics, and rapid proliferation [64, 65]. In addition to its role in regulation of oxidative stress, Nrf2 is also involved in the anabolic metabolism [65, 66**]. Nrf2 directly activates six genes involved in the PPP and nicotinamide adenine dinucleotide phosphate (NADPH) production pathway, including G6PD, PGD, TKT, TALDO1, and malicenzyme1 (ME1), through binding of this transcription factor to antioxidant response elements (AREs) of these gene promoters [66**]. Using [U-13C6] tracer assay, it has been demonstrated that Nrf2 is required for purine nucleotide synthesis [66**]. During metabolic reprogramming Nrf2 redirects glucose and glutamine into anabolic pathways, protecting cancer cells from oxidative damage [66**]. Activation of Nrf2 increases glucose uptake through the PPP, subsequently producing NADPH [66**–68**]. Our studies have reported that Nrf2 is constitutively activated in Cr(VI)-transformed cells and its inhibition suppresses tumorigenesis of these transformed cells [19]. Whether Nrf2 regulates PPP remains to be investigated in Cr(VI)-transformed cells. Our unpublished results indicate that Nrf2 positively regulates G6PD, PGD, TKT, and TALDO1 through direct binding to the AREs of these gene promoters, resulting in upregulation of PPP.
Glutaminolysis
Along with increased aerobic glycolysis, increased glutaminolysis is recognized as a key feature of the metabolic profile of cancer cells [69]. In addition to glycolysis, many tumors also depend on glutaminolysis to fuel their cellular bioenergetics and metabolism. Glutaminolysis catabolizes glutamine to downstream metabolites such as glutamate and α-ketoglutarate, important intermediates to fuel TCA cycle of tumors. Similar to glycolysis, glutaminolysis supplies cancer cells with both ATP and crucial precursors for continuous biosynthesis and accelerated proliferation [70, 71]. Table 1 shows that levels of three glutaminolytic enzymes including glutaminase (GLS), aspartate aminotransferase 2 (GOT2), and glutamine fructose-6-phosphate transaminase 1 (GFPT1) were elevated in Cr(VI)-transformed cells, suggesting upregulation of glutaminolysis.
Nrf2 increases glutamine consumption through enhancing glutaminolysis and glutathione synthesis. Nrf2 indirectly activates transcription factor 4 (ATF4), which regulates serine/glycine biosynthesis enzymes, supplying the substrates for glutathione and nucleotide production [72]. Nrf2 promotes glutathione synthesis from glutamine [37]. Nrf2 induces glutamate cysteine ligase (GCL), a key enzyme for glutathione synthesis, by directly activating the GCL encoding genes [73]. Nrf2 also increases the supply of cysteine by direct activation of the gene encoding cysteine transporter SLC7A11 [74]. Nrf2 is constitutively activated in Cr(VI)-transformed cells. The mechanism of Nrf2 in regulation of glutaminolysis in Cr(VI)-transformed cells has not yet been reported. In consideration of the findings from Table 1, it is very possible that Nrf2 targets GLS, which metabolizes glutamine to glutamate, providing a key nitrogen donor and carbon supply for the TCA cycle of Cr(VI)-transformed cells.
Conclusions and future perspectives
Metabolic reprogramming, a major hallmark of cancer, is characterized by upregulations of glycolysis, glutaminolysis, lipid metabolism, and pentose phosphate pathway. The metabolic reprogramming provides energy and metabolites to support rapid growth and proliferation of cancer cells. Chronic exposure of the cells to Cr(VI) causes malignant transformation. Similar to other cancer cells, these Cr(VI)-transformed cells have increased need for nutrients, energy, and biosynthetic activities to produce all macromolecular components during each passage through cell cycle. Cr(VI)-induced tumorigenesis is a chronic process. Among all studies related to bioenergetic phenotype induced by Cr(VI), most of studies focused on the short-term exposure. Only a few studies investigated the metabolic activities after long-term exposure to Cr(VI). For several decades, there has been a concentrated effort to identify the mechanisms of Cr(VI) carcinogenesis. However, little has been done to determine how changes of genes involved in glucose and glutamine metabolism contribute to Cr(VI) carcinogenesis. Cr(VI) exposure interferes with metabolic transduction pathways through different levels, including gene expression, intracellular protein levels, and protein function. We speculate that oxidative stress plays an important role in these processes. Chronic exposure of the cells to Cr(VI) causes ROS generation, leading to malignant cell transformation. Cr(VI)-transformed cells exhibit reduced capacity to generate ROS and elevated levels of antioxidant enzymes, contributing to development of apoptosis resistance and tumor growth. In Cr(VI)-transformed cells, PI3K/Akt/p38 signaling is activated, resulting in upregulation of glycolysis. Constitutive activation of Nrf2 in Cr(VI)-transformed cells enhances the PPP and NADPH generation, promoting cell proliferation. A representative scheme of possible mechanisms of metabolic reprogramming in Cr(VI) carcinogenesis is summarized in Figure 1. Many questions remain to be answered concerning the metabolic reprogramming in mechanism of carcinogenesis induced by Cr(VI) as well as other carcinogenic metals, such as arsenic, nickel, and cadmium. (a) Because Cr(VI) alters certain key proteins, which are important in metabolic reprogramming, it is important to understand whether certain metabolism-related pathways are the cause or the results of Cr(VI)-induced cells transformation and tumorigenesis. (b) It is also important to investigate whether and how metabolic reprogramming changes the cell process beyond energy metabolism, energy storage, and production of precursors for biosynthesis, which are major functions supporting malignancy and tumor growth. This study should cover the role of metabolic reprogramming in a broad area of Cr(VI) carcinogenesis, including cell cycle regulation , autophagy, malignant cell transformation, ROS generation, mitochondrial injury, apoptosis resistance, angiogenesis, migration, and invasion. (c) Many metabolic enzymes are regulated by allosteric binding or post-translational modification; for that reason, future studies should characterize metabolic fluxes to precisely evaluate the metabolic changes induced by Cr(VI). (d) It is also important to investigate whether interruption or alteration of metabolic reprogramming reduces Cr(VI)-induced malignant cells transformation and tumorigenesis. This study will lead to a new understanding on the mechanism of Cr(VI) carcinogenesis and its possible mechanism-based prevention.
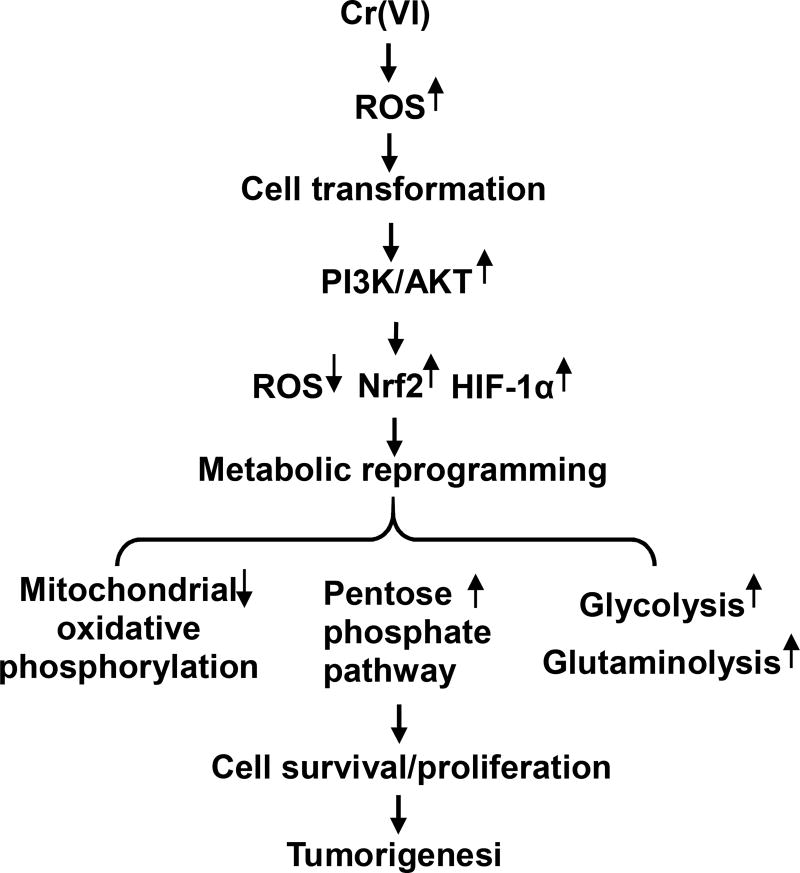
Figure 1. Representative scheme of metabolic reprogramming in Cr(VI) carcinogenesis.
Chronic exposure of cells to Cr(VI) causes ROS generation which is responsible for malignant cell transformation. Once the cells are malignant transformed, those cells exhibit activated PI3K/Akt, reduced ROS generation, and elevated levels of antioxidants and HIF-1α, resulting in reduction of mitochondrial oxidative phosphorylation and upregulations of pentose phosphate pathway, glycolysis, and glutaminolysis, leading to tumorigenesis.
Acknowledgments
This work is supported by National Institute of Health R01ES021771, R01ES025515, and R01ES025515S1 to Shi X and Zhang Z.
Abbreviations
- ACO1
Aconiatse
- ADPGK
ADP-specific glucokinase
- ALDOA
Aldolase A
- ARE
Antioxidant response element
- ATF4
Transcription factor 4
- CSCs
Cancer stem cells
- COX
Cytochrome C oxidase
- CS
Citrate synthase
- DLAT
Dihydrolipoamide S-acetyltransferase
- DLST
Dihydrolipoamide S-succinyltransferase
- ENO1
Enolase 1
- FBP1
Fructose-1,6-biosphosphatase
- FH
Fumarase
- G6PC3
Glucose-6-phosphatase 3
- G6PD
Glucose 6-phosphate dehydrogenase
- GCL
Glutamate cysteine ligase
- GFPT1
Glutamine-fructose-6-phosphate transaminase 1
- GLS
Glutaminase
- GOT2
Aspartate aminotransferase, mitochondrial
- GPI
Glucose-6-phosphate isomerase
- HIF
Hypoxia inducible factor
- HK2
Hexokinase
- MDH2
Malate dehydrogenase
- ME1
Malicenzyme1
- NDUF
NADH dehydrogenase
- NADPH:
Nicotinamide adenine dinucleotide phosphate
- Nrf2
Nuclear factor erythroid-(derived 2)-like 2
- OGDH
Oxoglutarate dehydrogenase
- OXPHOS
oxidative phosphorylation
- PDHA1
Pyruvate dehydrogenase E1
- PFKM
Phosphofructokinase
- PGD
6-phosphogluconate dehydrogenase
- PGK1
Phosphoglycerate kinase 1
- PKM
Pyruvate kinase
- PPP
Pentose phosphate pathway
- PRPS
Phosphorybosyl pyrophosphate synthetase
- RPE
Ribulose 5-phosphate 3-epimerase
- RPIA
Ribose 5-phosphate isomerase
- ROS
Reactive oxygen species
- SDHA
Succinate dehydrogenase
- SUCLG1
Succinyl-CoA synthase
- TALDO1
Transaldolase
- TKT
Transketolase
- UQCR
Ubiquinol cytochrome C reductase;
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
There are no conflicts of interest to declare.
References
(* of special interest and ** of outstanding interest)
- 1.Majumder PK, Febbo PG, Bikoff R, Berger R, Xue Q, McMahon LM, Golub TR, Loda M, Lane HA, Sellers WR. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat. Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 2.Shackelford DB, Vasquez DS, Corbeil J, Wu S, Leblanc M, Wu CL, Vera DR, Shaw RJ. mTOR and HIF-1alpha-mediated tumor metabolism in an LKB1 mouse model of Peutz-Jeghers syndrome. Proc. Natl. Acad. Sci. USA. 2009;106:11137–42. doi: 10.1073/pnas.0900465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stohs SJ, Bagchi D, Hassoun E, Bagchi M. Oxidative mechanisms in the toxicity of chromium and cadmium ions. J. Environ. Pathol. Toxicol. Oncol. 2000;19:201–13. [PubMed] [Google Scholar]
- 4.Ye J, Zhang X, Young HA, Mao Y, Shi X. Chromium(VI)-induced nuclear factor-κB activation in intact cells via free radical reactions. Carcinogenesis. 1995;16:2401–5. doi: 10.1093/carcin/16.10.2401. [DOI] [PubMed] [Google Scholar]
- 5.Yao H, Guo L, Jiang B, Luo J, Zhang Z, Shi X. Oxidative stress and chromium carcinogenesis. J. Environ. Pathol. Toxicol. Oncol. 2008;27:77–88. doi: 10.1615/jenvironpatholtoxicoloncol.v27.i2.10. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Son YO, Chang Q, Sun L, Hitron JA, Budhraja A, Zhang Z, Ke Z, Chen C, Luo J, Shi X. NADPH oxidase activation is required in reactive oxygen species generation and cell transformation induced by hexavalent chromium. Toxicol. Sci. 2011;123:399–410. doi: 10.1093/toxsci/kfr180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu S, Athar M, Lippai I, Waldren C, Hei T. Induction of oxyradicals by arsenic: implication for mechanism of genotoxicity. Proc. Natl. Acad. Sci. USA. 2001;98:1643–8. doi: 10.1073/pnas.031482998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barchowsky A, Klei LR, Dudek EJ, Swartz HM, James PE. Stimulation of reactive oxygen, but not reactive nitrogen species, in vascular endothelial cells exposed to low levels of arsenite. Free Rad. Biol. Med. 1999;27:1405–12. doi: 10.1016/s0891-5849(99)00186-0. [DOI] [PubMed] [Google Scholar]
- 9.Harris GK, Shi X. Signaling by carcinogenic metals and metal-induced reactive oxygen species. Mutation Res. 2003;533:183–200. doi: 10.1016/j.mrfmmm.2003.08.025. [DOI] [PubMed] [Google Scholar]
- 10.Machle W, Gregorius F. Cancer of the respiratory system in the United States chromate producing industry. Public Health Rep. 1948;63:1114–27. [PubMed] [Google Scholar]
- 11.Langard S. One hundred years of chromium and cancer: a review of epidemiological evidence and selected case reports. Am. J. Ind. Med. 1990;17:189–215. doi: 10.1002/ajim.4700170205. [DOI] [PubMed] [Google Scholar]
- 12.Langard S. Role of chemical species and exposure characteristics in cancer among persons occupationally exposed to chromium compounds. Scand. J. Work Environ. Health. 1993;19:81–9. [PubMed] [Google Scholar]
- 13.Agency for Toxic Substances and Disease Registry (ATSDR) Toxicological profile for chromium. U S Dep Health & Hum Serv. 1992 [PubMed] [Google Scholar]
- 14.Zhitkovich A. Chromium in drinking water: sources, metabolism, and cancer risks. Chem. Res. Toxicol. 2011;24:1617–1629. doi: 10.1021/tx200251t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhitkovich A. Importance of chromium-DNA adducts in mutagenicity and toxicity of chromium (VI) Chem. Res. Toxicol. 2005;18:3–11. doi: 10.1021/tx049774+. [DOI] [PubMed] [Google Scholar]
- 16.Woodruff TJ, Axelrad DA, Caldwell J, Morello-Frosch R, Rosenbaum A. Public health implications of 1990 air toxics concentrations across the United States. Environ. Health Perspect. 1998;106:245–51. doi: 10.1289/ehp.98106245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Shi X, Dalal NS. Chromium (V) and hydroxyl radical formation during the glutathione reductase-catalyzed reduction of chromium (VI) Biochem. Biophys. Res. Commun. 1989;163:627–34. doi: 10.1016/0006-291x(89)92183-9. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Pratheeshkumar P, Son YO, Kim D, Shi X. The role of reactive oxygen species in arsenic-induced transformation of human lung bronchial epithelial (BEAS-2B) cells. Biochem. Biophys. Res. Commun. 2015;456:643–8. doi: 10.1016/j.bbrc.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Son YO, Pratheeshkumar P, Wang Y, Kim D, Zhang Z, Shi X. Protection from Cr(VI)-induced malignant cell transformation and tumorigenesis of Cr(VI)-transformed cells by luteolin through Nrf2 signaling. Toxicol. Appl Pharmacol. 2017;331:24–32. doi: 10.1016/j.taap.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Dai J, Ji Y, Wang W, Kim D, Fai LY, Wang L, Luo J, Zhang Z. Loss of fructose-1,6-bisphosphatase induces glycolysis and promotes apoptosis resistance of cancer stem-like cells: an important role in hexavalent chromium-induced carcinogenesis. Toxicol. Appl. Pharmacol. 2017;15:164–173. doi: 10.1016/j.taap.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan J, Kamphorst JJ, Mathew R, Chung MK, White E, Shlomi T, Rabinowitz JD. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol. Syst. Biol. 2013;9:712. doi: 10.1038/msb.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, Lin Y, Yao J, Shi J, Kang T, Lorkiewicz P, St Clair D, Hung MC, Evers BM, Zhou BP. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23:316–31. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen M, Zhang J, Li N, Qian Z, Zhu M, Li Q, Zheng J, Wang X, Shi G. Promoter hypermethylation mediated downregulation of FBP1 in human hepatocellular carcinoma and colon cancer. PLoS One. 2011;6:e25564. doi: 10.1371/journal.pone.0025564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Wang J, Xing H, Li Q, Zhao Q, Li J. Down-regulation of FBP1 by ZEB1-mediated repression confers to growth and invasion in lung cancer cells. Mol. Cell. Biochem. 2016;411:331–40. doi: 10.1007/s11010-015-2595-8. [DOI] [PubMed] [Google Scholar]
- 25*.Li B, Qiu B, Lee DS, Walton ZE, Ochocki JD, Mathew LK, Mancuso A, Gade TP, Keith B, Nissim I, Simon MC. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251–5. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong BW, Qin GM, Luo Y, Mao JS. Metabolic enzymes: key modulators of functionality in cancer stem-like cells. Oncotarget. 2016;8:14251–67. doi: 10.18632/oncotarget.14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–15. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maxwell PH, Dachs GU, Gleadle JM, Nicholls LG, Harris AL, Stratford IJ, Hankinson O, Pugh CW, Ratcliffe PJ. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc. Natl. Acad. Sci. USA. 1997;94:8104–9. doi: 10.1073/pnas.94.15.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaupel P. The role of hypoxia-induced factors in tumor progression. Oncologist. 2004;9:10–7. doi: 10.1634/theoncologist.9-90005-10. [DOI] [PubMed] [Google Scholar]
- 30.Huss WJ, Hanrahan CF, Barrios RJ, Simons JW, Greenberg NM. Angiogenesis and prostate cancer: identification of a molecular progression switch. Cancer Res. 2001;61:2736–43. [PubMed] [Google Scholar]
- 31.Blancher C, Moore JW, Talks KL, Houlbrook S, Harris AL. Relationship of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression to vascular endothelial growth factor induction and hypoxia survival in human breast cancer cell lines. Cancer Res. 2000;60:7106–13. [PubMed] [Google Scholar]
- 32.Birner P, Schindl M, Obermair A, Plank C, Breitenecker G, Oberhuber G. Overexpression of hypoxia-inducible factor 1alpha is a marker for an unfavorable prognosis in early-stage invasive cervical cancer. Cancer Res. 2000;60:4693–6. [PubMed] [Google Scholar]
- 33.Giatromanolaki A, Koukourakis MI, Sivridis E, Turley H, Talks K, Pezzella F, Gatter KC, Harris AL. Relation of hypoxia inducible factor 1alpha and 2alpha in operable non-small cell lung cancer to angiogenic/molecular profile tumors and survival. Br. J. Cancer. 2001;85:881–90. doi: 10.1054/bjoc.2001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. Over-expression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5. [PubMed] [Google Scholar]
- 35.Kallio PJ, Wilson WJ, O’Brien S, Makino Y, Poellinger L. Regulation of the hypoxia-inducible factor 1alpha by the ubiquitin-proteasome pathway. J. Biol. Chem. 1999;274:6519–25. doi: 10.1074/jbc.274.10.6519. [DOI] [PubMed] [Google Scholar]
- 36.Huang LE, Arany Z, Livinston DM, Bunn HF. Activation of HIF depends primarily upon redox-sensitive stabilization of its alpha subunit. J. Biol. Chem. 1996;271:32253–9. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 37.Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat. Rev. Cancer. 2008;8:967–75. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 2010;20:51–6. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell. Metab. 2006;3:187–97. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell. Metab. 2006;3:177–85. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Kim D, Dai J, Park YH, Fai LY, Wang L, Pratheeshkumar P, Son YO, Kondo K, Xu M, Luo J, Shi X, Zhang Z. Activation of epidermal growth factor receptor/p38/Hypoxia-inducible factor-1α Is pivotal for angiogenesis and tumorigenesis of malignantly transformed cells induced by hexavalent chromium. J. Biol. Chem. 2016;291:16271–81. doi: 10.1074/jbc.M116.715797. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Kaczmarek M, Timofeeva OA, Karaczyn A, Malyguine A, Kasprzak KS, Salnikow K. The role of ascorbate in the modulation of HIF-1alpha protein and HIF-dependent transcription by chromium(VI) and nickel(II) Free Radic. Biol. Med. 2007;42:1246–57. doi: 10.1016/j.freeradbiomed.2007.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat. Rev. Cancer. 2008;8:51–6. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 44.Bensinger SJ, Christofk HR. New aspects of the Warburg effect in cancer cell biology. Semin. Cell Dev. Biol. 2012;23:352–61. doi: 10.1016/j.semcdb.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, Thompson CB. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–9. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 46.Robey RB, Hay N. Is Akt the Warburg kinase? - Akt-energy metabolism interactions and oncogenesis. Semin. Cancer Biol. 2009;19:25–31. doi: 10.1016/j.semcancer.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47*.Riganti C, Gazzano E, Polomeni A, Aldieri E, Ghigo D. The pentose phosphate pathway: an antioxidant defense and a crossroad in tumor cell fate. Free Radic. Biol. Med. 2012;53:421–36. doi: 10.1016/j.freeradbiomed.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 48.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jonas SK, Benedetto C, Flatman A, Hammond RH, Micheletti L, Riley C, Riley PA, Spargo DJ, Zonca M, Slater TF. Increased activity of 6-phosphogluconate dehydrogenase and glucose-6-phosphate dehydrogenase in purified cell suspensions and single cells from the uterine cervix in cervical intraepithelial neoplasia. Br. J. Cancer. 1992;66:185–91. doi: 10.1038/bjc.1992.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hartmannsberger D, Mack B, Eggert C, Denzel S, Stepp H, Betz CS, Gires O. Transketolase-like protein 1 confers resistance to serum withdrawal in vitro. Cancer Lett. 2011;300:20–9. doi: 10.1016/j.canlet.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 51.Koutras George A, Hattori Masao, Schneider Arthur S, Ebaugh Frank G, Jr, Valentine William N. Studies on Chromated Erythrocytes. Effect of Sodium Chromate on Erythrocyte Glutathione Reductase. J. Clin. Invest. 1964;43:323–31. doi: 10.1172/JCI104917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv. Enzyme Regul. 2006;46:113–40. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 53.Chan K, Han XD, Kan YW. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc. Natl. Acad. Sci. USA. 2002;98:4611–6. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54**.Sporn M, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat. Rev. 2012;12:564–71. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kostova AT. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid. Redox Signal. 2010;13:1713–48. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 56.Hu R, Saw CL, Yu R, Kong AN. Regulation of NF-E2-related factor 2 signaling for cancer chemo-prevention: antioxidant coupled with anti-inflammatory. Antioxid. Redox Signal. 2010;13:1679–98. doi: 10.1089/ars.2010.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57*.Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, Wong PK, Zhang DD. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–43. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL. The Keap1-Nrf2 pathway: mechanism of activation and dysregulatiion in cancer. Redox Biol. 2013;1:45–9. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohta T, Iijima K, Miyamoto M, NakaharaI I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, Shibata T, Yamamoto M, Hirohashi S. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–9. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 60.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol. Cell. Biol. 2006;26:2887–900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lau A, Whitman SA, Jaramillo MC, Zhang DD. Arsenic-mediated activation of the Nrf2-Keap1 antioxidant pathway. J. Biochem. Mol. Toxicol. 2013;27:99–105. doi: 10.1002/jbt.21463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, Moran CA, Corvalan AH, Biswal S, Swisher SG, Bekele BN, Minna JD, Stewart DJ, Wistuba II. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin. Cancer Res. 2010;16:3743–53. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. USA. 2008;105:13568–73. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, Wu H, Bova SG, Biswal S. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol. Cancer Ther. 2010;9:336–46. doi: 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, Ashush H, Coulter J, Blackford A, Goodman SN, Bunz F, Watson WH, Gabrielson E, Feinstein E, Biswal S. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–84. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66**.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 67**.Heiss EH, Schachner D, Zimmermann K, Dirsch VM. Glucose availability is a decisive factor for Nrf2-mediated gene expression. Redox Biol. 2013;1:359–65. doi: 10.1016/j.redox.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68**.Hawkins KE, Joy S, Delhove JM, Kotiadis VN, Fernandez E, Fitzpatrick LM, Whiteford JR, King PJ, Bolanos JP, Duchen MR, Waddington SN, McKay TR. NRF2 orchestrates the metabolic shift during induced pluripotent stem cell reprogramming. Cell Rep. 2016;14:1883–91. doi: 10.1016/j.celrep.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Daye D, Wellen KE. Metabolic reprogramming in cancer: unraveling the role of glutamine in tumorigenesis. Semin. Cell Dev. Biol. 2012;23:262–9. doi: 10.1016/j.semcdb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 70.Dang CV. Glutaminolysis: supplying carbon or nitrogen or both for cancer cells? Cell Cycle. 2010;9:3884–6. doi: 10.4161/cc.9.19.13302. [DOI] [PubMed] [Google Scholar]
- 71.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–24. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, Wistuba II, Minna JD, DeBerardinis RJ, Cantley LC. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 2015;47:1475–81. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sekhar KR, Crooks P, Sonar VN, Friedman DB, Chan JY, Meredith MJ, Starnes JH, Kelton KR, Summar SR, Sasi S, Freeman ML. NADPH oxidase activity is essential for Keap1/Nrf2-mediated induction of GCLC in response to 2-indol-3-yl-methylenequinuclidin-3-ols. Cancer Res. 2003;63:5636–45. [PubMed] [Google Scholar]
- 74.Sasaki H, Sato H, Kuriyama-Matsumura K, Sato K, Maebara K, Wang H, Tamba M, Itoh K, Yamamoto M, Bannai S. Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J. Biol. Chem. 2002;277:44765–71. doi: 10.1074/jbc.M208704200. [DOI] [PubMed] [Google Scholar]