Abstract
Human prostate cancer often metastasizes to bone, but the biological basis for such site-specific tropism remains largely unresolved. Recent work led us to hypothesize that this tropism may reflect pathogenic interactions between RAGE, a cell surface receptor expressed on malignant cells in advanced prostate cancer, and proteinase 3 (PR3), a serine protease present in inflammatory neutrophils and hematopoietic cells within the bone marrow microenvironment. In this study, we establish that RAGE-PR3 interaction mediates homing of prostate cancer cells to the bone marrow. PR3 bound to RAGE on the surface of prostate cancer cells in vitro, inducing tumor cell motility through a non-proteolytic signal transduction cascade involving activation and phosphorylation of ERK1/2 and JNK1. In preclinical models of experimental metastasis, ectopic expression of RAGE on human prostate cancer cells was sufficient to promote bone marrow homing within a short time frame. Our findings demonstrate how RAGE-PR3 interactions between human prostate cancer cells and the bone marrow microenvironment mediate bone metastasis during prostate cancer progression, with potential implications for prognosis and therapeutic intervention.
Keywords: bone metastasis, phage display, prostate cancer, RAGE, PR3
Introduction
Some prevalent tumors – prostate cancer among others – have selectivity for bone metastasis, but the biological basis for this observation remains poorly understood (1). Similar to hematopoietic stem cell homing to the bone marrow niche (2), it has been proposed that specific cell-cell and/or cell-matrix interactions might mediate tumor cell trafficking to the axial skeletal microenvironment. Accordingly, candidate protein interactions that promote osteoblastic proliferation of tumor and stromal cells have been identified (1). Understanding functional interactions between cancer cells and non-malignant cells within the bone marrow is central for developing target therapeutics for lethal metastatic tumors.
We have pioneered an approach to identify cancer-relevant ligand-receptors involving the selection of combinatorial random peptide libraries in terminal cancer patients (3,4). Using this strategy, we identified the peptide WELGGGP (single letter amino acid code) as targeting moiety for human bone marrow metastasis of prostate cancer (4). This motif mimics a portion of the ligand-binding domain of the receptor for advanced glycation end products (RAGE). The broad repertoire of RAGE ligands accumulates in tissues under cellular stress, contributing to chronic non-malignant diseases, such as diabetes and neurodegenerative disorders (5), and to malignant diseases, e.g. tumor cell activation during the metastatic cascade (6). Our analysis revealed that RAGE overexpression correlates with prostate cancer metastasis to bone marrow, but not to other non-osseous soft tissues such as lymph nodes (4). Moreover, proteinase 3 (PR3), a serine protease known to be secreted by cells of myeloid lineage (7), and allocated to the cell surface of neutrophils (8) and endothelial cells (9), was isolated as a RAGE-interacting protein (4). PR3 is the antigen of anti-neutrophil cytoplasmic antibodies that mediates inflammation with roles in disorders such as granulomatosis with polyangiitis (Wegener’s granulomatosis) (10). While a large number of studies provides evidence that PR3 contributes to inflammation, its role in cancer progression remains largely unknown.
Here, we analyzed the functional relevance of RAGE/PR3 interaction in the context of prostate cancer cell migration and bone marrow metastasis. Our results show that: (i) RAGE/PR3 binding mediates a heterotypic interaction between cancer cells and hematopoietic cells, (ii) binding of RAGE to PR3 activates a signaling cascade independent of PR3 proteolytic activity, which promotes tumor cell migration in vitro, and (iii) human prostate cancer cells expressing RAGE on their surfaces target the bone marrow microenvironment in vivo.
Materials and Methods
Cell lines
PC3, U937, HL-60 and DU145 cell lines were purchased from the American Type Culture Collection (ATCC) and were maintained in RPMI containing penicillin, streptomycin, L-glutamine, and 10% fetal bovine serum (FBS). pcDNA3.1(+)Nt-RAGE (N-terminal truncated) and pcDNA3.1(+)Fl-RAGE (full-length) plasmids were established in our laboratory between 2000 and 2004. Stably transfected PC3 cells were selected in complete culture containing 0.5 mg/mL G418 (Sigma). Validation of RAGE expression was analyzed by staining with a goat polyclonal (R&D System, PC3) or a mouse mAb (Abcam, DU145) anti-RAGE antibody at 200 ng/mL for 1 h at 4°C, followed by incubation with corresponding Cy3-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) at 5 μg/mL (30 min at 4°C) and flow cytometry analysis with a BD FACS Aria instrument (Becton Dickinson).
All cell lines were cytogenetically authenticated by ATCC, passaged 2–3 times, and stock aliquots were stored in liquid nitrogen. Cells were tested for Mycoplasma contamination by PCR and/or with the Universal Mycoplasma Detection Kit (ATCC, cat#30-1012K, http://www.atcc.org/products/all/30-1012K.aspx#generalinformation) or MycoAlert™ Mycoplasma Detection Kit (Lonza, Cat# LT07-318, http://www.lonza.com/products-services/bio-research/cell-culture-products/mycoplasma-detection-and-removal/mycoalert-mycoplasma-detection-kit.aspx). Thawed cells, passaged for <6 months, were not subjected to a new cell line authentication.
PR3 activity and cell motility assays
PR3 from primary human leukocytes (Sigma) was used unless otherwise specified. For proteolysis assays, 10 μg of recombinant proteins were incubated for 30 min at 37°C with 100 ng of PR3 prior to resolution by SDS-PAGE. For cell migration, 1×105 suspended cells were incubated for 30 min at 37°C with PR3 (0.3 μg/mL). After washing in PBS, cells were suspended in serum-free medium and applied to the upper chamber of transwell inserts (Costar) coated with Collagen 1 (Biosciences) in the presence of JNK inhibitor II (CAS 129-56-6 Calbiochem) (50 μM), or in control conditions. RPMI supplemented with 1% FBS was used as chemoattractant. After 24 h, migrated cells were fixed, stained in Crystal Violet (Fisher)-and counted (5 fields/well) under a light microscope.
Cell adhesion assays
Binding of PR3-expressing U937 cells to PC3 cells was assayed by co-culturing adherent PC3 cells with a ten-fold excess of U937 cells for 24 h, followed by washing with PBS. Binding of U937 and HL-60 cells to RAGE was assayed by coating 96-well plates with Fc-RAGE or control proteins (250 ng/well in 50 μl PBS), blocking with PBS containing 3% bovine serum albumin (BSA), followed by incubation with 5×104 U937, HL-60 or PC3 cells. In both cases, cells were fluorescently labeled with Calcein AM (Molecular Probes) in 150 mM NaCl and 20 mM HEPES, followed by washing and fluorescence quantification. Adherent cell numbers were normalized in a standard calibration curve. For cell adhesion inhibition assays, anti-RAGE antibody was used at 1.5 μg/mL.
Protein immunolocalization
Cells were grown overnight on coverslips in complete medium at 37°C followed by serum starvation for 16 hours. PR3 (0.5 μg/mL) was added to the cells for 30 min at 37°C. After washes, cells were fixed in 4% paraformaldehyde (PFA) for 10 min at room temperature (RT), permeabilized with 0.1% saponin, 0.1% BSA in TBS (30 min) and blocked with 1% BSA/TBS for 1 h at RT. Staining was performed with anti-RAGE antibody (Abcam) at 1 μg/mL, rabbit anti-PR3 antibody (Novus Biologicals) at 2 μg/mL, mouse monoclonal phospho-p44/42 (Thr202/Tyr204) E10 antibody (Cell Signaling) at 5 μg/mL, or rabbit anti-phospho-JNK (T183/Y185) antibody (R&D Systems) at 2 μg/mL. Secondary FITC-conjugated donkey anti-mouse antibody was used at 5 μg/mL and Cy3-conjugated goat anti-mouse or mouse anti-rabbit antibodies were used at 2.5 μg/mL (Jackson ImmunoResearch Laboratories). DNA was stained with Hoeschst 33258 (Sigma) or DAPI VectaShield (Vector Laboratories). Images were captured with an IX70 fluorescence microscope (Olympus) and confocal Leica TCS SP8.
Animal experiments
All experiments were approved by the Institutional Animal Care and Utilization Committees (IACUC) of University of Texas M. D. Anderson Cancer Center and University of New Mexico Health Science Center. Homing of PC3/Fl-RAGE, PC3/Nt-RAGE and DU145 to bone marrow was performed essentially as described (11). Briefly, cells were suspended in RPMI containing 5% FBS, and 5 μg of an anti-RAGE antibody was added to 100 μl of cell suspension and incubated for 1 h at RT prior to administration. Immunocompetent, C57BL/6 male mice (6-mo-old) were anesthetized with a tribromoethanol-based agent (Avertin; 0.015 mL/g/dose). The left cardiac ventricle was punctured percutaneously and 2×105 cells were administered through a 25-gauge needle attached to a 0.5 mL syringe. After 1 h, 10 mL of RPMI was perfused through the heart. Explanted tissues were fixed in 10% formalin, and bones were decalcified with 8% formic acid. Decalcification end point was determined by chemical test using a solution of 5% ammonium hydroxide and 5% ammonium oxalate.
Immunohistochemistry (IHC)
Immunostaining of formalin-fixed paraffin-embedded (FFPE) tissue sections was performed with mouse monoclonal pan-cytokeratin antibody (Abcam) used at 1:100 and LSAB+ peroxidase kit (DAKO). IHC staining of FFPE bone tissue was performed using the mouse monoclonal pan-cytokeratin antibody conjugated with HRP by Lightning-Link® Horseradish Peroxidase kit (Innova Bioscience) and revealed with DAB Chromogen kit (Biocare Medical). Hematoxylin was used as counterstain. Five random fields of each slide containing 5-μm tissue sections were imaged on a Nikon Ti Eclipse inverted microscope (Nikon Instruments). Cell count was determined using the Fiji ImageJ software. Positively stained cells were determined in Fiji using the Trainable Weka Segmentation plugin. Regions of positively stained-cells, non-stained cells and background in each field were marked and applied to train the classifier. Positive cells were determined from the final image constructed by the classifier.
Statistical analysis
GraphPad Prism software v.5.03 was used to graph data as mean ± standard error of the mean (SEM) and to calculate P-values with Student’s t-test (two-tailed) and two- way ANOVA. P<0.05 was considered statistically significant.
Results
Binding of RAGE to PR3 mediates heterotypic cell-cell adhesion
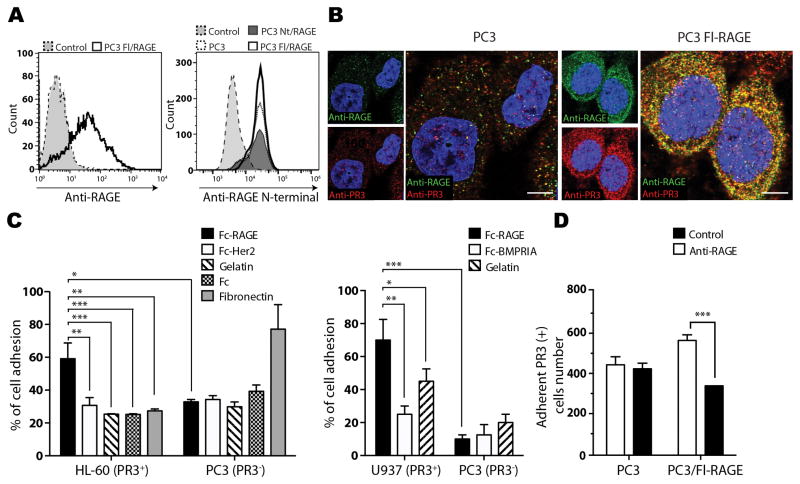
To test our hypothesis that prostate cancer cell-surface RAGE interacts with bone marrow-associated PR3, we first characterized the molecular basis of this interaction. To obtain in vitro models of RAGE-overexpressing or control cells, PC3 human prostate cancer cells, which express low levels of this receptor in culture (12), were stably transfected with full-length RAGE (PC3/Fl-RAGE) or with a N-truncated mutant lacking the WKLGGGP-spanning portion (PC3/Nt-RAGE) (13), respectively. RAGE expression and presence on the membrane were confirmed by flow cytometry analysis using anti-RAGE and anti-RAGE N-terminal antibodies (Fig. 1A). Addition of soluble PR3 to live cells resulted in accumulation at the cell surface in regions positive for RAGE, as evaluated by immunofluorescence of PC3 and PC3/Fl-RAGE cells (Fig. 1B). Intense colocalization of RAGE and PR3 was observed in PC3/Fl-RAGE cells when compared with control PC3. For in vitro binding assays, we used the human promyelocytic cell line HL-60 and myelomonocytic cell line U937, which both express PR3 (14). HL-60 and U937 cells specifically bound to wells coated with recombinant Fc fragment-fused RAGE (Fc-RAGE), but not with Fc alone, Fc-conjugated human epidermal growth factor 2 (Fc-Her2), Fc-conjugated bone morphogenic protein receptor 1A (Fc-BMPRIA) or gelatin. PC3 cells used as a negative control did not bind to Fc-RAGE or any of the control proteins, except fibronectin (Fig. 1C). Finally, an antibody against the WKLGGGP-spanning domain of human RAGE (4) inhibited binding of RAGE-expressing (PC3/Fl-RAGE) cells to PR3-expressing (U937) cells (Fig. 1D). Together, these results demonstrate a heterotypic cell-cell interaction between prostate cancer cells expressing RAGE and myeloid cells expressing PR3.
Figure 1.
PR3 binds to RAGE on prostate cancer cells. A, FACS analysis of cell surface full-length and N-terminal domain RAGE expression in PC3, PC3/Fl-RAGE and PC3/Nt-RAGE cells. White, white dashed line and dark grey: RAGE staining; grey: IgG isotype control. B, serum-starved PC3 and PC3/Fl-RAGE cells were incubated with 0.3 μg/mL PR3 for 30 min at 37°C, fixed and subjected to immunofluorescence with antibodies against RAGE (green) and PR3 (red). Co-localization is visualized as yellow signal. C, PR3-expressing (HL-60 and U937) or negative (PC3) cells were used in cell adhesion assays on immobilized Fc-RAGE or control proteins. Percentage of cell adhesion was calculated by relative fluorescence. D, binding of PR3-expressing U937 cells to RAGE-expressing cells is inhibited by anti-RAGE antibody. PC3/Fl-RAGE cells were pre-incubated with a control IgG or anti-RAGE antibody (1.5 μg/mL) and assayed for binding to suspended U937 cells. Error bars: SEM from three independent experiments. *P<0.05, ** P<0.01, *** P<0.001. Scale bar, 5 μm.
RAGE is a specific molecular interactor, but not a substrate, of PR3
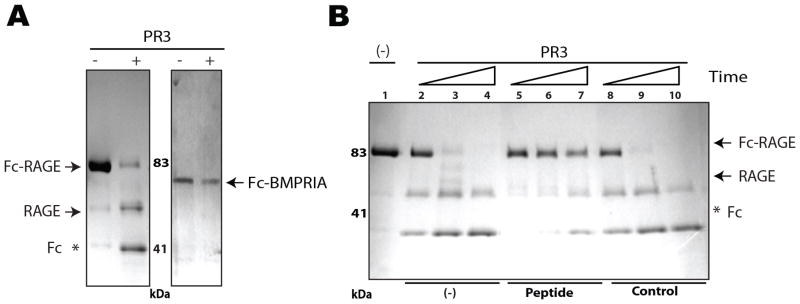
As PR3 is a proteinase, we analyzed whether RAGE could serve as a PR3 substrate. PR3 cleaved Fc-RAGE, but not another Fc-tagged recombinant protein, BMPRIA (Fig. 2A). Protein microsequencing revealed that this cleavage occurred not in the RAGE protein itself, but instead in the linker (amino acid sequence IEGRMD) that links RAGE to Fc. Because IEGRMD is the canonical sequence for Factor Xa, which is also a serine protease, we conclude that PR3 cleaves this linker as a result of binding to the RAGE moiety. Because Fc and BMPRIA are also linked by IEGRMD in the control protein, the lack of its cleavage indicates the specificity of PR3 binding to RAGE. This interaction between PR3 and RAGE was specifically inhibited by the peptide WELGGGP, as revealed by cleavage inhibition (Fig. 2B). Combined, these data are consistent with previous observation of RAGE binding to PR3 through the WKLGGGP motif located at the N-terminal region (4) and indicate that RAGE is not a substrate of PR3.
Figure 2.
Prostate cancer cell migration is induced by an enzyme activity-independent binding of PR3 to RAGE. A, Recombinant Fc-RAGE and Fc-BMPRIA (control) were exposed to PR3 (+) or mock buffer (−) for 1 h at 37°C, resolved by 4–20% SDS-PAGE and stained with Coomassie blue. (*) Fc portion cleaved off by PR3. B, Inhibition of Fc-RAGE proteolysis by RAGE-mimic peptide. Fc-RAGE (10 μg at 100 μg/mL) was incubated with 100 ng of PR3 (lanes 2–10) or buffer (lane 1) at 37°C for 1 min (lanes 2, 5, 8), 20 min (lanes 3, 6, 9), or 60 min (lanes 4, 7, 10) in the absence (lanes 2–4) or in the presence of 1 mg/mL biotinylated cyclic synthetic peptides: CWELGGGPC (lanes 5–7) or control (lanes 8–10), resolved by 4–20% SDS-PAGE and stained with Coomassie blue. Arrows indicate non-cleaved Fc-RAGE and cleaved RAGE. Fc portion is represented by (*).
PR3 activates MAP kinase pathway and cell motility in RAGE-expressing cells
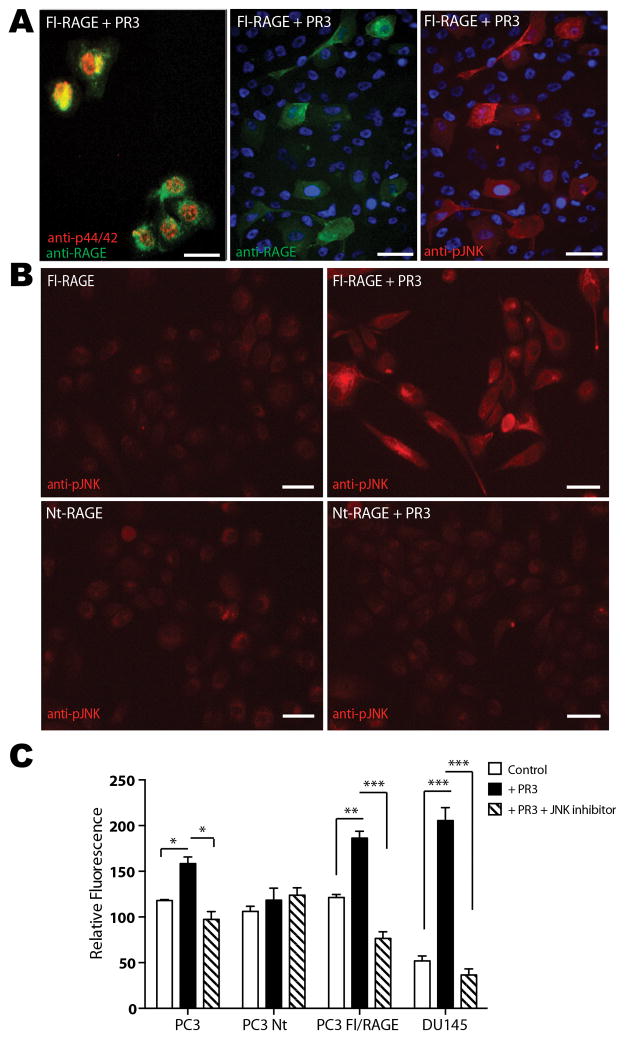
Binding of AGEs to RAGE promotes phosphorylation of p44/42 (Erk1/2) and c-Jun N-terminal kinase (JNK1), the signaling cascades activating cell motility (6,15). Due to the structural similarity of PR3 and advanced glycation end (AGE) products (4), we investigated whether PR3 binding to RAGE would trigger a similar signaling cascade and result in induction of tumor cell migration. We exposed PC3/Fl-RAGE cells to soluble PR3, and assayed MAP kinase phosphorylation status and localization by immunofluorescence. PC3/Fl-RAGE cells displayed nuclear phosphorylation of both p44/p42 (Fig. 3A) and JNK1 (Fig. 3A and B) upon PR3 treatment. Notably, high MAP kinase phosphorylation was detected in cells expressing high surface levels of RAGE (Fig. 3A). In contrast, no MAP kinase phosphorylation was observed in PC3/Nt-RAGE cells treated with soluble PR3 (Fig. 3B). Consistently with the activation of signal transduction pathways, we showed that PR3 significantly increased the motility of PC3, PC3/Fl-RAGE and DU145 cells, but not PC3/Nt-RAGE cells. Cell migration was inhibited in the presence of a JNK-specific inhibitor (Fig. 3C). Combined, these findings indicate that PR3 induces phosphorylation and nuclear translocation of p44/p42 and JNK1, leading to cancer cell motility, through a non-proteolytic interaction with cell-surface RAGE.
Figure 3.
PR3 activates RAGE-mediated MAP kinase phosphorylation. Where indicated, tumor cells were incubated in serum-free medium with 1 μg/mL PR3 for 30 min at 37°C prior to fixation. A, PC3/Fl-RAGE cells treated with PR3 were subjected to immunofluorescence with antibodies against the extracellular RAGE domain (green), p44/42 MAP kinase (red) or phosphorylated JNK1 (pJNK, red). Nuclei are stained blue. B, PC3/Fl-RAGE (top) or PC3/Nt-RAGE (bottom) cells with (right) or without (left) PR3 treatment and subjected to anti-pJNK immunofluorescence (red). Scale bar, 50 μm. C, RAGE/PR3 interaction induces tumor cell motility. Migration of PC3, PC3/Fl-RAGE, PC3/Nt-RAGE, and DU145 cells was evaluated in a transwell assay in the presence of PR3 and JNK inhibitor II, or in control conditions. The numbers of migrated cells are shown as mean ± SEM from three independent experiments. *P<0.05, ** P<0.01, *** P<0.001.
RAGE mediates homing of prostate cancer cells to the bone marrow
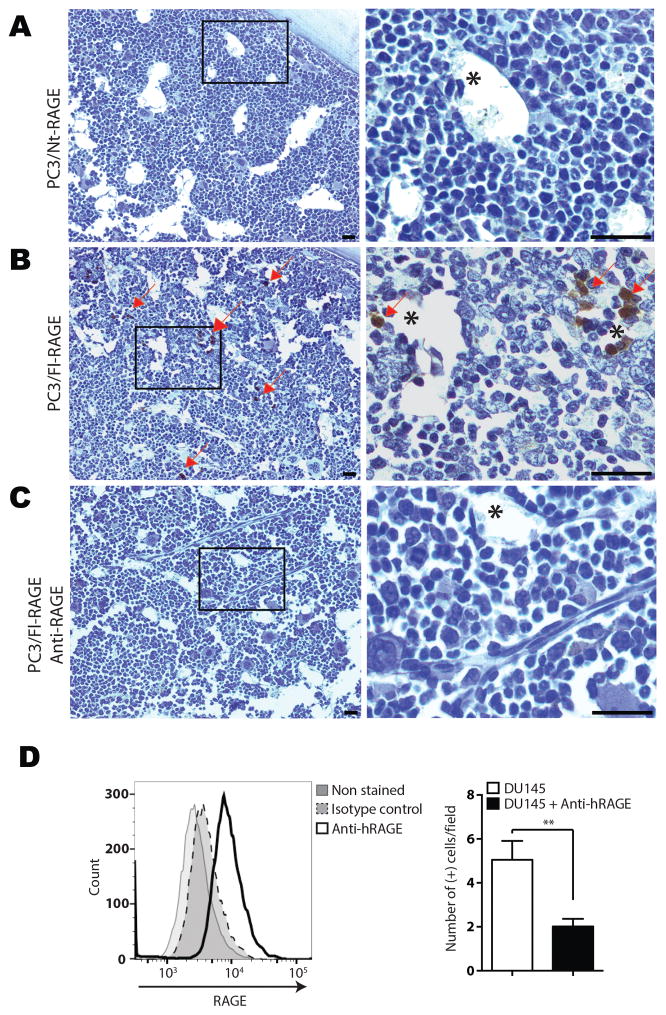
To assess the role of PR3/RAGE interaction in cancer metastasis, we evaluated whether RAGE would promote homing of circulating prostate cancer cells to the bone marrow. Based on the sequence similarity of mouse and human RAGE, we used human prostate cancer cells in immunocompetent mice. In our experimental model, we focused on homing of human prostate cancer cells that occurs within an hour prior to a species-specific immune response. Mice received intracardiac injection with either PC3/Nt-RAGE cells or PC3/Fl-RAGE in vehicle only or pre-incubated with anti-RAGE antibody. After 1 h, femur and tibia from both legs were harvested, along with control organs, fixed, and processed for histology. Decalcified bone sections were analyzed by staining for human cells with anti-human pan-cytokeratin antibody. IHC revealed abundant PC3/Fl-RAGE cells in bone marrow, where they appeared either as single cells or small clumps at the apical surface of capillaries (Fig. 4B). In contrast, PC3/Nt-RAGE cells were not detected in bone marrow microvasculature (Fig. 4A). Bone marrow homing of PC3/Fl-RAGE cells was significantly inhibited by pre-treatment with anti-RAGE blocking antibody (Fig. 4C). The same experiment was performed with human prostate cancer metastasis-derived DU145 cells, which express high levels of RAGE (12), with similar results (Fig. 4D). Combined, these data indicate that RAGE-expressing cancer cells interact with the bone marrow microenvironment, which is enriched in PR3-expressing cells.
Figure 4.
RAGE mediates bone marrow homing of prostate cancer cells. A, PC3/Nt-RAGE cells, B, PC3/Fl-RAGE cells, C, PC3/Fl-RAGE pre-incubated with human anti-RAGE antibody and D, DU145 cells. Control- or anti-RAGE antibody-treated cells were administered into the left heart ventricle of immunocompetent C57Bl/6 male mice. After 1 h, cardiac perfusion was performed, and tissues were removed and formalin-fixed. A–C, FFPE sections of bone marrow from formic acid-decalcified femur bones were processed for IHC with anti-human pan-cytokeratin HRP-conjugated antibodies. High magnification of insets is shown on the right. Arrows: PC3/Fl-RAGE cells (brown). Asterisks: blood vessels. Hematoxylin counter-staining is shown in blue. Scale bar, 50 μm. D, Expression of RAGE in DU145 cells was determined by flow cytometry using human anti-RAGE antibody. The IHC quantification of cells stained positive with the anti-human anti-pan cytokeratin antibody (right) was performed using Trainable Weka Segmentation plugin from Fiji Image J software. ** P<0.01.
Discussion
Our group demonstrated that it is possible to proceed rapidly from discovery in humans (3) to preclinical validation (16,17) to a first-in-human clinical trial (18). From four ligand-receptors identified in terminal patient, two are ubiquitous and previously reported (integrin α4/annexinA4 and cathepsin B/apolipoprotein E3) and the other two are specific to white adipose tissue (WAT) (prohibitin/annexin A2) (19) and tumor tissues (RAGE/PR3) (4), respectively. In prostate cancer, high level of RAGE was observed in patients with metastatic disease, and specifically in bone (but not lymph node) metastases (4). The clinical relevance of this observation has remained unclear.
Here we present ex vivo and in vivo evidence that binding of PR3 to RAGE, followed by activation of p44/42 and JNK1 in prostate cancer cells, induces cell motility and tumor homing to the bone marrow. RAGE/PR3 interaction is likely important at two steps of metastasis: (i) cancer cell mobilization from the primary tumor and (ii) cancer cell homing and attachment to the bone marrow. Supporting a role in the first step is a well-recognized association of cancer progression with inflammation induced by PR3 expressed by leukocytes within the tumor microenvironment (7). Moreover, activation of p44/p42 and JNK1 in primary tumor cells has been shown to induce matrix metalloproteinases and angiogenic factors responsible for increased tumor invasiveness (6,15). Thus, RAGE signaling pathways likely facilitate the initial stage of tumor cell dissemination. With respect to the second step, our data indicate that the interaction of RAGE with PR3 mediates adhesion of circulating prostate tumor cells within the bone marrow. In this context, PR3 on promyeloid progenitors and/or sinusoid endothelial cells could serve as “soil” for prostate cancer homing. Finally, PR3 could promote re-activation of MAP kinase pathways in cancer cells forming micrometastases, resulting in their extravasation.
Several ligands have been reported to interact with RAGE and trigger activation of signaling pathways related to cellular migration, proliferation, and survival (5). Tumor invasiveness and metastatic potential have been correlated with RAGE upregulation, and blocking RAGE-ligand interactions has been shown to suppress tumor progression (15). It is probable that in cancers not primarily predisposed to bone metastasis RAGE mediates tumor cell homing through RAGE-binding proteins (known or as yet unknown) other than PR3, and with different functional attributes. Future studies will be required to further characterize the organ-specific heterotypic interactions that could be investigated as targets of prospective anti-metastasis therapies.
Précis.
Interactions between a prostate cancer cell surface receptor and a proteinase expressed by myeloid cells in the bone microenvironment are found to drive the most common form of metastasis during prostate cancer progression, with immediate implications for molecular prognosis and intervention.
Acknowledgments
We thank Dr. Thiruvengadam Arumugam and Dr. Craig D. Logsdon (University of Texas M. D. Anderson Cancer Center) for the pcDNA3.1(+)Nt-RAGE and pcDNA3.1(+)Fl-RAGE plasmids (13). Images in this paper were generated in the University of New Mexico & Cancer Center Fluorescence Microscopy Shared Resource, funded as detailed on: http://hsc.unm.edu/crtc/microscopy/acknowledgement.shtml.
Financial Support: This work was supported by grants from the National Institutes of Health (P50CA100632 and P01CA148600 to J.J. Molldrem, Cancer Center Support Grant P30CA016672 to MD Anderson Cancer Center) and the US Department of Defense Prostate Cancer Research Program (W. Arap), and by awards from AngelWorks, the Gillson Longenbaugh Foundation, the Marcus Foundation, and the Prostate Cancer Foundation (R. Pasqualini and W. Arap).
Footnotes
Conflict of interest statement: R.P. and W.A. are founders of and equity holders in Alvos Therapeutics. R.P. and W.A. are inventors listed on patent applications related to this work and will be entitled to standard royalties if licensing and/or commercialization occurs. The University of New Mexico Health Sciences Center currently manages these arrangements in accordance with its established institutional conflict of interest policy.
References
- 1.Sturge J, Caley MP, Waxman J. Bone metastasis in prostate cancer: emerging therapeutic strategies. Nat Rev Clin Oncol. 2011;8(6):357–68. doi: 10.1038/nrclinonc.2011.67. [DOI] [PubMed] [Google Scholar]
- 2.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505(7483):327–34. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arap W, Kolonin MG, Trepel M, Lahdenranta J, Cardó-Vila M, Giordano RJ, et al. Steps toward mapping the human vasculature by phage display. Nat Med. 2002;8(2):121–7. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]
- 4.Staquicini FI, Cardó-Vila M, Kolonin MG, Trepel M, Edwards JK, Nunes DN, et al. Vascular ligand-receptor mapping by direct combinatorial selection in cancer patients. Proc Natl Acad Sci U S A. 2011;108(46):18637–42. doi: 10.1073/pnas.1114503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierhaus A, Humpert PM, Morcos M, Wendt T, Chavakis T, Arnold B, et al. Understanding RAGE, the receptor for advanced glycation end products. J Mol Med (Berl) 2005;83(11):876–86. doi: 10.1007/s00109-005-0688-7. [DOI] [PubMed] [Google Scholar]
- 6.Kalea AZ, See F, Harja E, Arriero M, Schmidt AM, Hudson BI. Alternatively spliced RAGEv1 inhibits tumorigenesis through suppression of JNK signaling. Cancer research. 2010;70(13):5628–38. doi: 10.1158/0008-5472.CAN-10-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alatrash G, Mittendorf EA, Sergeeva A, Sukhumalchandra P, Qiao N, Zhang M, et al. Broad cross-presentation of the hematopoietically derived PR1 antigen on solid tumors leads to susceptibility to PR1-targeted immunotherapy. J Immunol. 2012;189(11):5476–84. doi: 10.4049/jimmunol.1201221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Csernok E, Ernst M, Schmitt W, Bainton DF, Gross WL. Activated neutrophils express proteinase 3 on their plasma membrane in vitro and in vivo. Clin Exp Immunol. 1994;95(2):244–50. doi: 10.1111/j.1365-2249.1994.tb06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayet WJ, Csernok E, Szymkowiak C, Gross WL, Meyer zum Buschenfelde KH. Human endothelial cells express proteinase 3, the target antigen of anticytoplasmic antibodies in Wegener’s granulomatosis. Blood. 1993;82(4):1221–9. [PubMed] [Google Scholar]
- 10.Gabillet J, Millet A, Pederzoli-Ribeil M, Tacnet-Delorme P, Guillevin L, Mouthon L, et al. Proteinase 3, the autoantigen in granulomatosis with polyangiitis, associates with calreticulin on apoptotic neutrophils, impairs macrophage phagocytosis, and promotes inflammation. J Immunol. 2012;189(5):2574–83. doi: 10.4049/jimmunol.1200600. [DOI] [PubMed] [Google Scholar]
- 11.Sun YX, Schneider A, Jung Y, Wang J, Dai J, Wang J, et al. Skeletal localization and neutralization of the SDF-1(CXCL12)/CXCR4 axis blocks prostate cancer metastasis and growth in osseous sites in vivo. J Bone Miner Res. 2005;20(2):318–29. doi: 10.1359/JBMR.041109. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto KI, Murata H, Putranto EW, Kataoka K, Motoyama A, Hibino T, et al. DOCK7 is a critical regulator of the RAGE-Cdc42 signaling axis that induces formation of dendritic pseudopodia in human cancer cells. Oncol Rep. 2013;29(3):1073–79. doi: 10.3892/or.2012.2191. [DOI] [PubMed] [Google Scholar]
- 13.Arumugam T, Simeone DM, Schmidt AM, Logsdon CD. S100P stimulates cell proliferation and survival via receptor for activated glycation end products (RAGE) J Biol Chem. 2004;279(7):5059–65. doi: 10.1074/jbc.M310124200. [DOI] [PubMed] [Google Scholar]
- 14.Rao NV, Rao GV, Marshall BC, Hoidal JR. Biosynthesis and processing of proteinase 3 in U937 cells - Processing pathways are distinct from those of cathepsin G. Journal of Biological Chemistry. 1996;271(6):2972–78. doi: 10.1074/jbc.271.6.2972. [DOI] [PubMed] [Google Scholar]
- 15.Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, Qu W, et al. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature. 2000;405(6784):354–60. doi: 10.1038/35012626. [DOI] [PubMed] [Google Scholar]
- 16.Zurita AJ, Troncoso P, Cardó-Vila M, Logothetis CJ, Pasqualini R, Arap W. Combinatorial screenings in patients: the interleukin-11 receptor alpha as a candidate target in the progression of human prostate cancer. Cancer research. 2004;64(2):435–9. doi: 10.1158/0008-5472.can-03-2675. [DOI] [PubMed] [Google Scholar]
- 17.Cardó-Vila M, Marchio S, Sato M, Staquicini FI, Smith TL, Bronk JK, et al. Interleukin-11 Receptor Is a Candidate Target for Ligand-Directed Therapy in Lung Cancer Analysis of Clinical Samples and BMTP-11 Preclinical Activity. Am J Pathol. 2016;186(8):2162–70. doi: 10.1016/j.ajpath.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasqualini R, Millikan RE, Christianson DR, Cardó-Vila M, Driessen WH, Giordano RJ, et al. Targeting the interleukin-11 receptor alpha in metastatic prostate cancer: A first-in-man study. Cancer. 2015;121(14):2411–21. doi: 10.1002/cncr.29344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salameh A, Daquinag AC, Staquicini DI, An Z, Hajjar KA, Pasqualini R, et al. Prohibitin/annexin 2 interaction regulates fatty acid transport in adipose tissue. JCI Insight. 2016;1(10) doi: 10.1172/jci.insight.86351. [DOI] [PMC free article] [PubMed] [Google Scholar]