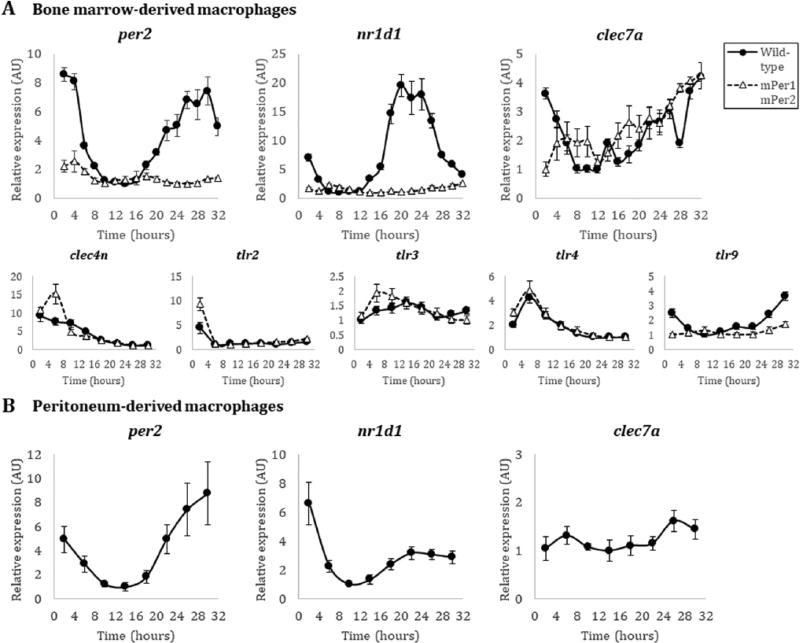
Figure 1.
Transcript expression of fungal-recognition receptor genes in macrophages. (A) Quantitative PCR of per2, nr1d1, clec7a, clec4n, tlr2, tlr3, tlr4, and tlr9 transcripts from male wild-type mPer2Luc/Luc (black filled circle, solid line) and male mutant mPer1m/mmPer2m/m (white filled triangle, dashed line) BMDMs in the C57BL/6 background. (B) Quantitative PCR of per2, nr1d1, and clec7a transcripts from wild-type mPer2Luc/Luc peritoneum-derived macrophages in the C57BL/6 background. Time (hours) represents time after initial serum synchronization. Error bars represent ± SE from technical replicates, n=3. Methods. Because the 129 strain of mice used to create the knock-outs/knock-ins (Yoo et al., 2004; Zheng et al., 2001) is not isogenic with the C57BL/6J background, and to avoid subtle genetic background issues, we carried out crosses to the C57BL/6J background, followed by SNP mapping, to select the best progeny to speed the track to isogenicity with C57. After repeated backcrosses, mPer2Luc/Luc mice used here are >97% homozygous with the C57BL/6J background.