Abstract
African Americans (AAs) who have PCa typically have more aggressive disease and make up a disproportionate number of the disease deaths, relative to European Americans (EAs). TMPRSS2 translocations, a common event in EA patients, are exploited in diagnostic and prognostic settings, whereas they are diminished in frequency in AA men. Thus, these patients with TMPRSS2 fusion-negative disease represent an under-investigated patient group. We propose that epigenetic events are a significant and alternative driver of aggressive disease in fusion-negative PCa. To reveal epigenetically governed microRNAs (miRNAs) that are enriched in fusion-negative disease and associated with aggressive in AA PCa, we leveraged both our experimental evidence and publically available data. These analyses identified 18 miRNAs that are differentially altered in fusion-negative disease, associated with DNA CpG methylation, and implicated in aggressive and AA PCas. Understanding the relationships between miRNA expression, upstream epigenetic regulation by DNA methylation, and downstream regulation of mRNA targets in fusion negative disease is imperative to understanding the biological basis of the racial health disparity in PCa.
Keywords: miRNA, DNA Methylation, TMPRSS2:ERG, Prostate Cancer, African American, Review
2. GENETIC AND EPIGENETIC DRIVERS OF PROSTATE CANCER (PCa) ARE INTERTWINED
Amongst men in the US, PCa is most common non-cutaneous cancer diagnosed and second leading cause of death (1, 2). This cancer is heterogeneous in terms of progression rates, and various genetic and epigenetic alterations are prevalent and appear to be factors in the tumorigenesis and progression of PCa.
Currently, in PCa, the clinical parameters do not accurately predict progression risks to more aggressive stages of disease. For example, the parameters used for assigning men to radical prostatectomy (PSA level, TNM stage and Gleason score) lack the specificity and sensitivity to distinguish accurately men who both need and will be cured by this treatment, from those who will experience treatment failure typified with rising PSA following surgery (so-called biochemical failure) (3, 4). This is of clinical significance, as patients who experience treatment failure are more likely to progress to more aggressive forms of PCa with increased risks of tumor-related mortality (5, 6). Furthermore, compounding these uncertainties are issues concerning the sensitivity and specificity of the PSA test itself (4). The result of this ambiguity is that men who do not require therapy are treated with either radical surgery or high-dose radiation because their physicians are unable to tell who “needs” therapy (1, 2). Thus, identifying and characterizing non-invasive biomarkers to identify those men who need more aggressive treatment are required.
This ambiguity is further obscured because the incidence and natural history of PCa varies between races. American men of African ancestry have a 19% higher incidence and 37% higher mortality from PCa compared to men of European ancestry (reviewed in (7–10)). Thus, in African American (AA) PCa patients, the disease appears more aggressive and occurs at a younger age, than in European American (EA) patients.
In an effort to define the disease, multiple groups, including the Cancer Genome Atlas (TCGA) consortium, have added to previous understanding (11, 12) and have established roles for common genetic alterations in PCa (13–15) and novel somatic mutations, including Mediator of RNA Polymerase II Transcription, Subunit 12 (MED12), Forkhead Box A1 (FOXA1), and Speckle-Type POZ Protein (SPOP). The E3 ubiquitin ligase adaptor, SPOP, shows recurrent mutations in 6–15% of tumors across multiple independent cohorts (14, 16). Also supporting the importance of androgen receptor (AR) signaling in PCa and the cross–talk with epigenetic events, the coactivator Nuclear Receptor Coactivator 2 (NCOA2) is commonly amplified (15, 17). Furthermore, mapping of copy number variations re-enforces the significance of PTEN loss and MYC gain (18). Recently, genetic changes have been used to map clonal evolution of metastatic PCa tumors and have revealed the possibility of multi-cancer clonal evolution within a single tumor (19, 20).
The complex nature of cancer phenotypes, however, cannot be explained by genetic components alone (21). Epigenomic modifications and events contribute to cell transformation. These events can be defined as heritable changes to the expression and regulation of gene expression that are not associated with alteration in the DNA sequences (22), although of course deregulated epigenetic events are often downstream of genetic changes (23, 24).
Epigenomic events include the control of chromatin structure and the function of non-coding RNAs, including microRNA (miRNA). Chromatin structure, which is dynamically regulated and generates a highly plastic interface between transcription factors and the genome, governs gene expression. Similarly, non-coding RNA species affect both chromatin structure and transcriptional responses and regulate transcriptional networks. Therefore, these events impact many aspects of cell signaling and function (reviewed in (25)). In turn, chromatin structure is regulated by hundreds of proteins, often acting in large complexes, that initiate local and large-scale control of the position of nucleosomes and the post-translational modifications to the histones contained within nucleosomes (26–29) (reviewed in (30)). These actions also integrate with other complexes that govern the status of DNA CpG methylation. In this manner, the epigenome regulates the transcriptional control of gene networks related to cell proliferation, differentiation, and programmed death.
Epigenomic events appear to play distinct yet complementary roles to genomic events and add to an explanation for the basis of cancer, for many cancers show gene-specific and global changes in DNA CpG methylation and/or altered histone modification patterns (30–34). For example, up-regulation of the histone methyltransferase, Enhancer of Zeste Homolog 2 (EZH2), appears commonly in both localized and metastatic PCas and associates with a poorer prognosis (35, 36). These histone modifications govern tissue-specific gene regulation and are often intertwined with the actions of DNA methyltransferases (DNMTs) (37) to bring about stable and heritable changes in the capacity (both positive and negative) for regulation of gene expression.
3. MICRORNA ACT AS EPIGENETIC DRIVERS OF PCa
In parallel to the well-established understanding on the role of classical epigenetic events, the role of non-coding RNA in epigenetic regulation of gene expression has also emerged. The four types of small RNAs, namely, small nuclear (sn)RNAs, small nucleolar (sno)RNAs, miRNAs, and transfer (t)RNAs, correspond to as much as 85% of total small annotated RNAs as determined by GENCODE. Similarly, ENCODE data reveals the wide range expression of miRNAs in different normal and cancer cell lines (38), supporting the concept that miRNAs are components of gene regulation networks and are expressed differentially in disease stages. Although miRNAs represent only ~1% of the genome, they are estimated to target 30% of the genes (39). Furthermore, there is de-regulation of miRNAs for a variety of solid tumors, including breast cancers, colon cancers, and PCas (40–44).
In PCa, several miRNAs are differentially regulated and act as both tumor suppressors and onco-miRs. For instance, miR-221 and miR-222 are upregulated in castration-resistant PCa cells (45) and appear to control cyclin-dependent kinase (Cdk) inhibitors p27KIP1 and p57KIP2, and thus the cell cycle (46, 47), PI3K and PTEN signaling (48), and a range of other signaling events (49–53), although some contradictory findings suggest that the subtleties of miRNA function are not yet fully revealed (54).
Similarly, miR-125b and miR-143 are upregulated in serum samples from patients with metastatic PCa as compared to those from normal individuals (55). Expression of miR-125b in serum of PCa patients is reported to be upregulated as compared to normal controls (56); other studies report it to be downregulated in PCa as compared to normal or benign prostatic hyperplastic samples (57–59). miR-125b regulates cell proliferation in PCa cell lines (60), and is suggested to be upregulated by androgen signaling (61). Functionally, in PCa, miR-125b targets BAK1 (61) (a pro-apoptotic member of the BCL-2 gene family) and EIF4EBP1 (Eukaryotic translation initiation factor 4E-binding protein 1), a gene that encodes a member of a family of translation repressor proteins (58).
Other miRNAs have been identified as tumor suppressors, including miR-143, miR-145, and the miR-200 family. Expression of miR-143 and miR-145 are suppressed in PCas and negatively associate with metastasis (62). These miRs contribute to PCa progression though the epithelial-mesenchymal transition (EMT) (62) and loss of their repressive effect on the EGFR/RAS/MAPK pathway (62). miR-205 and miR-200 family miRNAs, also downregulated in PCas, regulate the EMT by targeting ZEB1 and ZEB2 in PCas (50, 63, 64).
4. EXPLOITING SERUM EXPRESSION OF miRNA TO PREDICT PCa
Understanding and exploiting serum miRNA classifiers in PCa patients offers an alternative route to accurate diagnosis of disease. Mandel and Metais first identified serum-borne nucleic acids in 1948; of these, miRNAs potentially hold the greatest diagnostic promise (reviewed in (65)). miRNAs appear to contribute to multiple oncogenic actions in PCas (reviewed in (58). They are secreted into serum, where they remain stable (66–69) and can be reliably extracted and measured (56, 70–72). Using serum-borne molecules as prognostic markers is attractive for several reasons. First, they can overcome the limitations of inaccurate sampling of the prostate gland for the presence of cancer. Second, they can encapsulate the effects of heterotypic cell interactions within the tumor micro-environment. Third, they can be used as a non-invasive test procedure and, therefore, hold considerable promise to be exploited as accurate and functional prognostic markers of PCa. Reflecting this potential, studies have now established that miRNA expression can identify PCa that is at an advanced stage and distinguish it from less aggressive or indolent states, and tumors that have the potential to become invasive from non-invasive, organ-confined tumors (56–58, 73–77).
Expression of miRNAs is frequently disrupted in malignancies (reviewed in (42)), and epigenetic inactivation via promoter CpG island hypermethylation is commonly observed for miRNAs with putative growth-inhibitory functions (reviewed in (78)). For example, in PCa, promoter hypermethylation is associated with loss of miR-200 family members that regulate the EMT and cell migration/invasion. Similarly, hypermethylation of the promoter of miR-34a prevents its regulation by p53 and thereby distorts apoptosis and other functions (79–82). However, to date, most studies have focused on DNA methylation and miRNA interactions in general without asking if these alterations differ in either specific subgroups or racial populations.
These functional relationships and mechanisms of disruption suggest that miRNAs can be exploited to reveal the basis for fusion-negative PCas. Furthermore, miRNAs hold considerable promise to be exploited as accurate and functional prognostic serum markers of PCa (83–87) because they can encapsulate events within the tumor micro-environment and overcome the limitations of inaccurate tumor sampling at biopsy. From a biostatistical perspective, given that there are fewer miRNAs than mRNAs, genome-wide coverage is more readily achieved, and the statistical penalties typically associated with mRNA genome-wide testing can be avoided (88).
5. TRANSLOCATIONS OF THE TMPRRS2 GENE ASSOCIATE WITH AGGRESSIVE DISEASE BUT DIFFER IN THEIR INCIDENCE ACROSS RACES
To identify men with aggressive cancer, Chinnaiyan and colleagues pioneered approaches for identifying and exploiting translocations across the genome in PCa (89–94). This group identified common, and unsuspected, translocations of the TMPRSS2 gene and the ETS-Related Gene (ERG), which is part of the ETS transcription factor family that has oncogenic functions. The TMPRSS2 gene codes for an androgen-responsive protease specific to prostate cells, whose expression and genetic variation is associated with aggressive PCa (95, 96). Therefore, this fusion leads to overexpression of ERG, which in turn regulates multiple genes, including AR-responsive genes (91) (reviewed (97)). Following this discovery (94), and as a result of these pivotal studies, much attention has been given to the TMPRSS2–ERG and related ETS family translocations as potential biomarkers, with over 600 publications following. These gene translocations are common and are related to disease stage and progression risks (98–101). There is therefore good evidence that they act as an androgen-activated tumor driver, justifying clinical exploitation; however, the biological and clinical significance of this event remains an active research area (102–108). As a result, there have been efforts to exploit detection of these translocation products in urine. The goal of these studies has been to generate a clinically approved, urine-based approach to aid in early detection, in the so-called Mi-Prostate Score (89).
This clinical development depends on frequency of the translocation products in the patient population. The TMPRSS2–ERG translocation (94) is commonly and consistently identified in approximately 50% of EAs. This focus has to some extent overlooked the fact that the prevalence of these fusion products is lower for other ethnicities including AA, Japanese, and Chinese patients (109). The rate of TMPRSS2–ERG translocations in AA men appears to be significantly lower, in the range of 10–30% (110–113). AA patients, in addition to having lower incidence of TMPRSS2:ERG fusion, also have lower expression of ERG protein (109, 114, 115). For example, in matched cohorts of 91 AA and 91 EA men, there was lower expression of ERG in AA vs EA index tumors (29% vs 63%) (116). This was more pronounced for higher-grade cancers, in which nearly 60% of EA patients were ERG-positive, whereas only 10% of AA patients were ERG-positive (117, 118). Reflecting this discrepancy, the Mi-Prostate Score test is more accurate for EA than AA patients (119). A follow-up publication to these findings found that, in 154 AA men compared to 243 EA men, AA men are more likely to be negative for many common translocations including: ERG-negative, ETS-negative, SPINK1-negative disease (51% v 35%; p=0. 0. 02) (5, 120). The absence of these translocations correlated with the AA PCa disease. Thus, while the TMPRSS:ERG fusion may help to diagnose PCa in EA patients, its low frequency in AA patients suggests that it neither helps to explain disease etiology nor aid with accurate diagnosis of aggressive disease in this group. In fact, this presents a problem for all patients who are fusion-negative, but it is particularly devastating for men of African ancestry.
Thus, dissecting the biological basis for TMPRSS2:ERG fusion-negative PCa represents an underexplored and clinically significant need for AA patients and has the potential to be exploited to define new diagnosis and treatment options. To meet this need, we have explored the role of miRNA-mediated mechanisms that drive disease in TMPRSS2 fusion-negative PCas. The rationale was that miRNAs act as alternative epigenetic drivers of PCa and allow precise stratification of patients. The goal was to develop a biological understanding of tumor drivers in AA men.
6. ARE miRNA RACE-SPECIFIC, EPIGENETIC DRIVERS FOR TMPRSS2 FUSION-NEGATIVE PCa?
Across and within races, the biological basis for TMPRSS2 fusion-negative tumors remains unexplored. Our recent findings and the work of others, however, suggests that epigenetic drivers are consequential in this form of PCa. There are racial differences in the methylome of AA and EA PCa patients (121–124). For example, AR, RARβ2, and other genes have higher methylation prevalence in tumors of AA patients, whereas TIMP3 has higher methylation in non-involved normal AA prostate tissues (125). DNA methylation has also emerged as a regulator of expression for various miRNAs in the progression in PCa (126–128), but the extent of differential CpG methylation in AA and EA PCa remains under-explored.
There are global differences in the pattern of CpG DNA methylation in benign, cancerous, and metastatic samples in fusion-positive vs fusion-negative PCas (129). Similarly, in a larger cohort, methylated DNA immunoprecipitation sequencing identified differential CpG methylation based on the TMPRSS2–ERG rearrangement status (130). These data suggest that DNA methylation events distort gene expression patterns in fusion-negative PCa and drive this disease. Although these events are significant across the genome, we have pursued the relationships between DNA methylation and miRNA expression for strategic reasons.
Together, these data support the concept that altered methylation of miRNA provides an alternative route for the initiation and progression of PCa in AA patients, whose tumors are commonly TMPSS2 fusion-negative. This raises the possibility that racially specific hypermethylation of key miRNAs drives alternative routes for the initiation and progression of PCa in AA patients, independent of TMPRSS2:ERG fusion.
7. AN ANALYTICAL PIPELINE TO EXAMINE CHANGES IN miRNA IN TMPSS2 FUSION-NEGATIVE PCa
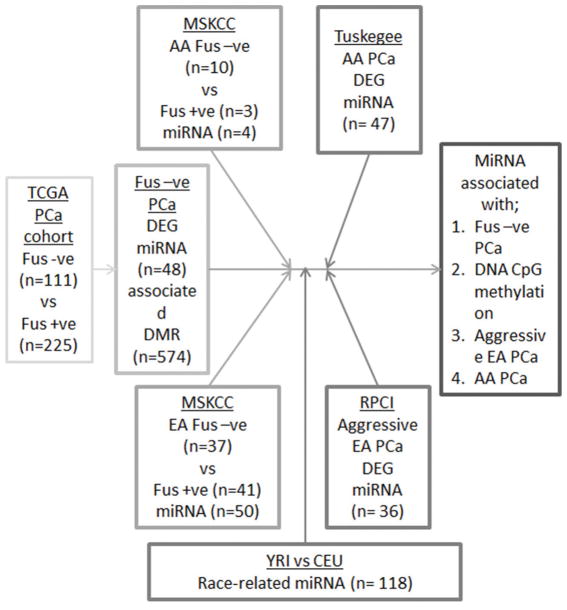
To test this broad goal, we developed an integrative genomic pipeline to identify miRNAs that are associated with altered DNA methylation and TMPRSS2 fusion-negative PCas. We subsequently integrated our previous findings on miRNAs that are predictive of aggressive tumors and AA PCas (86, 87) and combined these findings (17, 131, 132) to identify, in fusion-negative PCa, which of these miRNAs is altered through DNA methylation (Figure 1).
Figure 1.
An integrative pipeline to identify miRNA altered by DNA methylation in TMPRSS2 fusion negative PCa. Publically available data from the main TCGA cohort tumors (15) and the Memorial Sloan Kettering Cancer Center (MSKCC) cohort of 91 tumors (17) was examined to identify miRNA that differentially expressed and associated with differential DNA methylation. Subsequently these miRNAs were examined for being differentially expressed between TMPRSS2 fusion positive and negative tumors, expression in lymphoblastoid cells from 53 CEU (Utah residents with northern and western European ancestry) and 54 YRI (Yoruba people from Ibadan, Nigeria) populations, and known from our previous studies to be associated with aggressive PCa, and enriched in AA PCa.
Specifically, we performed analyses of differential expression of miRNA genes (DEG) in publically available data by mining two different PCa cohorts in TCGA: the main TCGA cohort of localized PCa (15) and the Memorial Sloan Kettering Cancer Center (MSKCC) cohort of 91 tumors (17). In the main cohort, we identified 48 miRNA DEG in altered fusion-negative vs -positive disease (pcorrected < 0. 0. 5) that also associated with reciprocal and CpG DNA methylation (pcorrected < 0. 0. 5).
In the MSKCC cohort, we identified miRNA DEG in fusion-negative vs -positive disease in smaller numbers of AA and EA patients. To this, we also combined miRNA expression associated with race. Huang et al. (133) evaluated differences in population-level expression in 757 miRNAs in HapMap lymphoblastoid cell lines derived from 53 CEU (Utah residents with northern and western European ancestry) and 54 YRI (Yoruba people from Ibadan, Nigeria). Of all miRNAs evaluated, and after stringent correction for multiple testing, 33 differed between these two ethnic groups, pcorrected < 0. 0. 5.
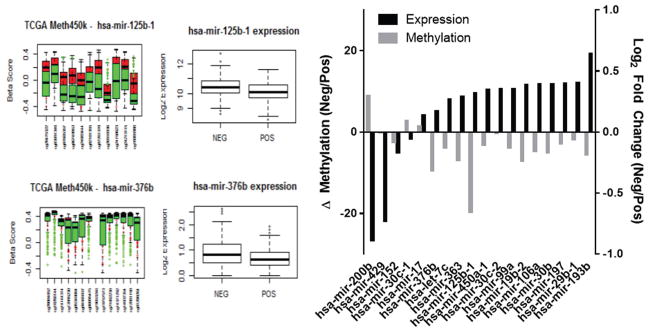
Finally, these miRNAs were filtered further to focus on miRNAs that we had established to be associated with disease progression, and with AA PCas identified in patient samples and cell lines from Roswell Park Cancer Institute (RPCI) (86) and Tuskegee University (TU) (87). This approach identified 18 miRNAs as associated with aggressive PCa. These are candidates to drive alternative progression pathways in fusion-negative PCas in AA and EA patients. Across these miRNAs, there was an inverse correlation between CpG methylation and expression of associated TMPRSS2 fusion-negative disease, thus providing proof-of-concept that the race-specific miRNAs are differentially expressed in TMPRSS2:ERG-negative tumors (Figure 2). Some of these miRNAs (e. g. miR-125b) have been investigated extensively in prostate tissue, whereas others have few, if any, publications related to PCa (Table 1). For example, miR-376b, which was initially identified from the TU cohort and verified in the YRI cohort to be ancestry-related, has a loss of methylated CpG islands in the promoter region in TMPRSS2:ERG-negative tumors compared to TMPRSS2:ERG-positive tumors; this pattern is seen for many of the miRNAs. miR-376b is associated with the mTOR-related autophagy proteins, TG4C and BECN1, and relates to angiogenesis through regulation of the HIF1alpha VEGFA/Notch1 signaling pathway (134, 135).
Figure 2.
Inverse relationships between CpG methylation and expression of 18 miRNAs associated with TMPRSS2 fusion-negative disease across 333 localized tumors in the TCGA cohort (15) (ρ = −0. 5. 6).
Table 1.
Summary of miRNA associated with TMPRSS2 fusion-negative PCa and inversely correlated with DNA CpG methylation
| miRNA | Family | Clustered | Serum Detected | Number of PCa pubs |
|---|---|---|---|---|
| Let-7c | Let-7 | mir-99a | Y | 9 |
| miR-106a | Mir-17 | mir-19b, mir-363 | Y* | 7 |
| miR-125b1 | Mir-10 | Y | 22 | |
| miR-152 | Mir-148 | Y | 5 | |
| miR-17 | Mir-17 | mir-19b | Y* | 17 |
| miR-19b | Mir-19 | mir-17 | Y | 2 |
| miR-193b | Mir-193 | Y | 3 | |
| miR-197 | Mir-197 | Y | 1 | |
| miR-29b1 | Mir-29 | Y | 10 | |
| miR-200b | Mir-8 | mir-429 | Y | 13 |
| miR-30b | Mir-30 | Y | 1 | |
| miR-30c1 | Mir-30 | Y | 5 | |
| miR-30c2 | Mir-30 | N | 5 | |
| miR-363 | Mir-363 | mir106a, mir-19b | Y | 1 |
| miR-376b | Mir368 | Y | - | |
| miR-429 | Mir-8 | mir-200b | Y | 1 |
| miR-450 | Mir-450 | Y | - | |
| miR-99a | Mir-10 | Let-7c | Y | 6 |
The 18 miRNAs represent 13 families, and several cluster < 10 kb from each other. A pilot study was undertaken to examine miRNA expression in the sera from 8 EA PCa patients and 7 AA PCa patients. 17 of 18 miRNAs were detectable in the sera and two were significantly different (*) between the two groups. Some of these miRNAs are well-described in PCas (e. g. , miR-125b), whereas others are unexplored (e. g. , miR-376b).
8. DOWNSTREAM TARGETS OF KEY miRNA
For several of these miRNAs, we have also characterized known targets and suggest that there is convergence on regulators of the epigenome. For example, miR-125b targets Nuclear Receptor Corepressor 2/Silencing Mediator Of Retinoic Acid and Thyroid Hormone Receptor (NCOR2/SMRT) (136) and an interacting co-factor that governs CpG methylation dependent silencing, Zinc Finger And BTB Domain Containing 33 (ZBTB33/Kaiso), which targets miR-125a-5p, a member of the miR-125 family (137). Reflecting changes in miR-125b expression in patients who experience biochemical failure, we separately revealed, in a cohort of 172 PCa patients, that Kaiso is overexpressed in a cohort of AA PCa patients in the University of Alabama at Birmingham/TU cohort (138). As determined with another tissue microarray of 720 PCa patients from RPCI, NCOR2/SMRT expression also predicts progression risk. Similarly, miR-152 appears to regulate proteins involved in control of the epigenome. Ectopic expression of miR-152 results in decreased cellular proliferation, migration, and invasion, and decreased expression of DNMT1 through binding in the DNMT1 3’UTR (87) and thereby contributes to aberration of hypermethylation in AA tumors (87). Thus, our approaches support the concept that disease progression is influenced by miRNA in both EA and AA tumors and that increased aggressiveness of AA tumors may reflect both common and unique targetomes of mRNA networks. In particular, our studies have suggested that, within these targetomes, are mRNAs that encode for epigenetic regulators of DNA methylation and histone modifications. This suggests a complex interplay between upstream epigenetic regulation and downstream regulation of epigenetic regulators, which together determines disease risk.
In the AA PCa cohort of miRNAs, we have examined the impact of DNA methylation on some of the miRNAs. Validation of specific miRNAs by q-PCR across a panel of 11 cell lines demonstrated that miR-132, miR-367b, miR-410, and miR-152 were decreased in the more aggressive PCa cells, and expression of each miRNA was reversed after treatment of the cells with 5-aza-2′-deoxycytidine, suggesting a role for CpG methylation. Others have suggested that, in PCas, miR-132 is silenced by methylation (139). Our data confirm that miR-152 is silenced by methylation in PCas (87). In a comparison of normal/tumor ratios of miR-152 expression in 20 EA and 20 AA PCa patients, miR-152 expression was decreased in tumors of 67% of all patients, with 50 % of the AA patients showing a difference, compared to only 35% of EA patients. We also found that miR-152 expression was lower in non-malignant tissues from AA patients compared to those of EAs, suggesting the presence of heritable differences in the control of miR-152 expression.
The impact of genetic variation on miRNA expression. In support of these heritable differences, three single nucleotide polymorphisms (SNPs) on chromosome 17 (rs1553754, rs11079828, and rs6504340) are associated with miR-152 expression at the genome-wide level, and differ in minor allele frequency by 28% on average in comparing the YRI and CEU populations (140, 141). In fact, the minor alleles at two of these SNPs for miRNA expression quantitative trait loci (miR-eQTL), rs1553754 and rs6504340, are different in the YRI and CEU populations. These differences in the frequency of alleles associated with miRNA expression are evidence that the difference in methylation rates in miR-152 could be driven by differences in genetic variation seen between African and European populations. In addition to this population-level evidence, through our work, we have identified rs12940701 (C/T) in miR152; the minor allele (T) has a relatively high frequency (26%%) in the CEU population versus the YRI population (11%), suggesting that EAs have lower methylation rates due to the presence of a T nucleotide instead of a C nucleotide, which is less frequent in the Yoruba population. Indeed, the number of publications that demonstrate an increased rate of methylation in AAs compared to EAs is growing, supporting the value of these miRNAs as diagnostic for AA PCa.
9. PERSPECTIVES
There is an urgent need to identify and characterize minimally-invasive diagnostic markers of PCa progression in TMPRSS2:ERG fusion-negative AA patients, as these patients are generally underserved. By use of AA cell cultures and publicly available datasets, we have undertaken a discovery phase of interrogating miRNAs that associate with aggressive PCa. Several fundamental questions remain as to the impact of methylation on silencing of miRNA expression in PCas, particularly in AA patients. Thus, validation of the capability of these miRNA serum biomarkers to predict aggressive PCa in AAs is required before these miRNAs can be considered for clinical application.
In preliminary studies, we have examined expression of these 18 miRNAs in the sera of EA and AA PCa patients with matched stage and grade of disease. In this pilot study, only one miRNA (miR-30c-5p) was undetected in PCa patient serum. Even within this small set of patients, however, several miRNAs were expressed at different levels in AA vs EA patients.
These findings suggest that epigenetic events affected by genetic variation differentially regulate miRNAs in AA PCa patients and are drivers of TMPRSS2:ERG-negative tumors. Identification of differentially methylated regions in AA and EA PCas will aid in defining a more personalized risk assessment of developing aggressive PCa, for not only AA men, but all men with TMPRSS2:ERG-negative tumors.
Acknowledgments
This work was supported by the Prostate Program of the Department of Defense Congressionally Directed Medical Research Programs (W81XWH-14-1-0608, W81XWH-11-2-0033 to MJC and PC120913 to CY). MJC and LESC acknowledge support, in part, of the National Cancer Institute Cancer Center Support Grant to the Roswell Park Cancer Institute (CA016056). CY also acknowledges support from the National Institute for Health, specifically G12 RR03059-21A1 (NIH/NIMHD), 1 R21 CA188799-01 (NIH/NCI), and U54 CA118623 (NIH/ NCI).
References
- 1.Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, Denis LJ, Recker F, Berenguer A, Maattanen L, Bangma CH, Aus G, Villers A, Rebillard X, van der Kwast T, Blijenberg BG, Moss SM, de Koning HJ, Auvinen A. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360(13):1320–8. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 2.Andriole GL, Crawford ED, Grubb RL, 3rd, Buys SS, Chia D, Church TR, Fouad MN, Gelmann EP, Kvale PA, Reding DJ, Weissfeld JL, Yokochi LA, O’Brien B, Clapp JD, Rathmell JM, Riley TL, Hayes RB, Kramer BS, Izmirlian G, Miller AB, Pinsky PF, Prorok PC, Gohagan JK, Berg CD, Team PP. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360(13):1310–9. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freedland SJ. Screening, risk assessment, and the approach to therapy in patients with prostate cancer. Cancer. 2011;117(6):1123–35. doi: 10.1002/cncr.25477. [DOI] [PubMed] [Google Scholar]
- 4.Cookson MS, Aus G, Burnett AL, Canby-Hagino ED, D’Amico AV, Dmochowski RR, Eton DT, Forman JD, Goldenberg SL, Hernandez J, Higano CS, Kraus SR, Moul JW, Tangen C, Thrasher JB, Thompson I. Variation in the definition of biochemical recurrence in patients treated for localized prostate cancer: the American Urological Association Prostate Guidelines for Localized Prostate Cancer Update Panel report and recommendations for a standard in the reporting of surgical outcomes. The Journal of urology. 2007;177(2):540–5. doi: 10.1016/j.juro.2006.10.097. [DOI] [PubMed] [Google Scholar]
- 5.Yamoah K, Deville C, Vapiwala N, Spangler E, Zeigler-Johnson CM, Malkowicz B, Lee DI, Kattan M, Dicker AP, Rebbeck TR. African American men with low-grade prostate cancer have increased disease recurrence after prostatectomy compared with Caucasian men. Urol Oncol. 2015;33(2):70 e15–22. doi: 10.1016/j.urolonc.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ludwig MS, Kuban DA, Strom SS, Du XL, Lopez DS, Yamal JM. Assessing the Optimum Use of Androgen-Deprivation Therapy in High-Risk Prostate Cancer Patients Undergoing External Beam Radiation Therapy. Am J Mens Health. 2015 doi: 10.1177/1557988315581396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones J, Grizzle W, Wang H, Yates C. MicroRNAs that affect prostate cancer: emphasis on prostate cancer in African Americans. Biotech Histochem. 2013;88(7):410–24. doi: 10.3109/10520295.2013.807069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powell IJ, Bollig-Fischer A. Minireview: the molecular and genomic basis for prostate cancer health disparities. Mol Endocrinol. 2013;27(6):879–91. doi: 10.1210/me.2013-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drake BF, Keane TE, Mosley CM, Adams SA, Elder KT, Modayil MV, Ureda JR, Hebert JR. Prostate cancer disparities in South Carolina: early detection, special programs, and descriptive epidemiology. J S C Med Assoc. 2006;102(7):241–9. [PubMed] [Google Scholar]
- 10.Cooney KA. Hereditary prostate cancer in African-American families. Semin Urol Oncol. 1998;16(4):202–6. [PubMed] [Google Scholar]
- 11.Li PE, Nelson PS. Prostate cancer genomics. Curr Urol Rep. 2001;2(1):70–8. doi: 10.1007/s11934-001-0028-6. [DOI] [PubMed] [Google Scholar]
- 12.Isaacs WB. Molecular genetics of prostate cancer. Cancer Surv. 1995;25:357–79. [PubMed] [Google Scholar]
- 13.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, Van Allen E, Kryukov GV, Sboner A, Theurillat JP, Soong TD, Nickerson E, Auclair D, Tewari A, Beltran H, Onofrio RC, Boysen G, Guiducci C, Barbieri CE, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Ramos AH, Winckler W, Cipicchio M, Ardlie K, Kantoff PW, Berger MF, Gabriel SB, Golub TR, Meyerson M, Lander ES, Elemento O, Getz G, Demichelis F, Rubin MA, Garraway LA. Punctuated evolution of prostate cancer genomes. Cell. 2013;153(3):666–77. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, Nickerson E, Chae SS, Boysen G, Auclair D, Onofrio RC, Park K, Kitabayashi N, MacDonald TY, Sheikh K, Vuong T, Guiducci C, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Hussain WM, Ramos AH, Winckler W, Redman MC, Ardlie K, Tewari AK, Mosquera JM, Rupp N, Wild PJ, Moch H, Morrissey C, Nelson PS, Kantoff PW, Gabriel SB, Golub TR, Meyerson M, Lander ES, Getz G, Rubin MA, Garraway LA. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.s. c. m. o. Cancer Genome Atlas Research Network. Electronic address and N. Cancer Genome Atlas Research. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163(4):1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, Nickerson E, Chae SS, Boysen G, Auclair D, Onofrio RC, Park K, Kitabayashi N, MacDonald TY, Sheikh K, Vuong T, Guiducci C, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Hussain WM, Ramos AH, Winckler W, Redman MC, Ardlie K, Tewari AK, Mosquera JM, Rupp N, Wild PJ, Moch H, Morrissey C, Nelson PS, Kantoff PW, Gabriel SB, Golub TR, Meyerson M, Lander ES, Getz G, Rubin MA, Garraway LA. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature genetics. 2012;44(6):685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hieronymus H, Schultz N, Gopalan A, Carver BS, Chang MT, Xiao Y, Heguy A, Huberman K, Bernstein M, Assel M, Murali R, Vickers A, Scardino PT, Sander C, Reuter V, Taylor BS, Sawyers CL. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A. 2014;111(30):11139–44. doi: 10.1073/pnas.1411446111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, Brewer DS, Kallio HM, Hognas G, Annala M, Kivinummi K, Goody V, Latimer C, O’Meara S, Dawson KJ, Isaacs W, Emmert-Buck MR, Nykter M, Foster C, Kote-Jarai Z, Easton D, Whitaker HC, Group IPU, Neal DE, Cooper CS, Eeles RA, Visakorpi T, Campbell PJ, McDermott U, Wedge DC, Bova GS. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520(7547):353–7. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper CS, Eeles R, Wedge DC, Van Loo P, Gundem G, Alexandrov LB, Kremeyer B, Butler A, Lynch AG, Camacho N, Massie CE, Kay J, Luxton HJ, Edwards S, Kote-Jarai Z, Dennis N, Merson S, Leongamornlert D, Zamora J, Corbishley C, Thomas S, Nik-Zainal S, Ramakrishna M, O’Meara S, Matthews L, Clark J, Hurst R, Mithen R, Bristow RG, Boutros PC, Fraser M, Cooke S, Raine K, Jones D, Menzies A, Stebbings L, Hinton J, Teague J, McLaren S, Mudie L, Hardy C, Anderson E, Joseph O, Goody V, Robinson B, Maddison M, Gamble S, Greenman C, Berney D, Hazell S, Livni N, Group IP, Fisher C, Ogden C, Kumar P, Thompson A, Woodhouse C, Nicol D, Mayer E, Dudderidge T, Shah NC, Gnanapragasam V, Voet T, Campbell P, Futreal A, Easton D, Warren AY, Foster CS, Stratton MR, Whitaker HC, McDermott U, Brewer DS, Neal DE. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015;47(4):367–72. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev. 2012;22(1):50–5. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Holliday R. The inheritance of epigenetic defects. Science. 1987;238(4824):163–70. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- 23.Valdes-Mora F, Clark SJ. Prostate cancer epigenetic biomarkers: next-generation technologies. Oncogene. 2015;34(13):1609–18. doi: 10.1038/onc.2014.111. [DOI] [PubMed] [Google Scholar]
- 24.Cooper CS, Foster CS. Concepts of epigenetics in prostate cancer development. Br J Cancer. 2009;100(2):240–5. doi: 10.1038/sj.bjc.6604771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16(3):178–89. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 26.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nature reviews. Molecular cell biology. 2007;8(4):284–95. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 27.Kim HJ, Bae SC. Histone deacetylase inhibitors: molecular mechanisms of action and clinical trials as anti-cancer drugs. American journal of translational research. 2011;3(2):166–79. [PMC free article] [PubMed] [Google Scholar]
- 28.Albert M, Helin K. Histone methyltransferases in cancer. Seminars in cell & developmental biology. 2010;21(2):209–20. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 29.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nature reviews. Molecular cell biology. 2012;13(5):297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 30.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286–98. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 31.Campbell MJ, Turner BM. Altered histone modifications in cancer. Advances in experimental medicine and biology. 2013;754:81–107. doi: 10.1007/978-1-4419-9967-2_4. [DOI] [PubMed] [Google Scholar]
- 32.Chiam K, Ricciardelli C, Bianco-Miotto T. Epigenetic biomarkers in prostate cancer: Current and future uses. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 33.Jeronimo C, Bastian PJ, Bjartell A, Carbone GM, Catto JW, Clark SJ, Henrique R, Nelson WG, Shariat SF. Epigenetics in prostate cancer: biologic and clinical relevance. Eur Urol. 2011;60(4):753–66. doi: 10.1016/j.eururo.2011.06.035. [DOI] [PubMed] [Google Scholar]
- 34.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24(18):1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 36.Saramaki OR, Tammela TL, Martikainen PM, Vessella RL, Visakorpi T. The gene for polycomb group protein enhancer of zeste homolog 2 (EZH2) is amplified in late-stage prostate cancer. Genes Chromosomes Cancer. 2006;45(7):639–45. doi: 10.1002/gcc.20327. [DOI] [PubMed] [Google Scholar]
- 37.Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO reports. 2011;12(7):647–56. doi: 10.1038/embor.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489(7414):101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis BP, Burge CB, BarTel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 40.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. Acomprehensive review. EMBO molecular medicine. 2012;4(3):143–59. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manikandan J, Aarthi JJ, Kumar SD, Pushparaj PN. Oncomirs: the potential role of non-coding microRNAs in understanding cancer. Bioinformation. 2008;2(8):330–4. doi: 10.6026/97320630002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature reviews. Cancer. 2006;6(4):259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 43.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 45.Sun T, Wang Q, Balk S, Brown M, Lee GS, Kantoff P. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009;69(8):3356–63. doi: 10.1158/0008-5472.CAN-08-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, Croce CM, Stein GS. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer research. 2008;68(8):2773–80. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mercatelli N, Coppola V, Bonci D, Miele F, Costantini A, Guadagnoli M, Bonanno E, Muto G, Frajese GV, De Maria R, Spagnoli LG, Farace MG, Ciafre SA. The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice. PLoS ONE. 2008;3(12):e4029. doi: 10.1371/journal.pone.0004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Tang H, Thayanithy V, Subramanian S, Oberg AL, Cunningham JM, Cerhan JR, Steer CJ, Thibodeau SN. Gene networks and microRNAs implicated in aggressive prostate cancer. Cancer research. 2009;69(24):9490–7. doi: 10.1158/0008-5472.CAN-09-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hao J, Zhang C, Zhang A, Wang K, Jia Z, Wang G, Han L, Kang C, Pu P. miR-221/222 is the regulator of Cx43 expression in human glioblastoma cells. Oncol Rep. 2012;27(5):1504–10. doi: 10.3892/or.2012.1652. [DOI] [PubMed] [Google Scholar]
- 50.Tucci P, Agostini M, Grespi F, Markert EK, Terrinoni A, Vousden KH, Muller PA, Dotsch V, Kehrloesser S, Sayan BS, Giaccone G, Lowe SW, Takahashi N, Vandenabeele P, Knight RA, Levine AJ, Melino G. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(38):15312–7. doi: 10.1073/pnas.1110977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283(45):31079–86. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283(44):29897–903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin D, Cui F, Bu Q, Yan C. The expression and clinical significance of GTP-binding RAS-like 3 (ARHI) and microRNA 221 and 222 in prostate cancer. J Int Med Res. 2011;39(5):1870–5. doi: 10.1177/147323001103900530. [DOI] [PubMed] [Google Scholar]
- 54.Wach S, Nolte E, Szczyrba J, Stohr R, Hartmann A, Orntoft T, Dyrskjot L, Eltze E, Wieland W, Keck B, Ekici AB, Grasser F, Wullich B. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. International journal of cancer. Journal international du cancer. 2012;130(3):611–21. doi: 10.1002/ijc.26064. [DOI] [PubMed] [Google Scholar]
- 55.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(30):10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tong AW, Fulgham P, Jay C, Chen P, Khalil I, Liu S, Senzer N, Eklund AC, Han J, Nemunaitis J. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009;16(3):206–16. doi: 10.1038/cgt.2008.77. [DOI] [PubMed] [Google Scholar]
- 58.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27(12):1788–93. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 59.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer research. 2007;67(13):6130–5. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 60.Lee YS, Kim HK, Chung S, Kim KS, Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem. 2005;280(17):16635–41. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- 61.Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, Tepper CG, Evans CP, Kung HJ, deVere White RW. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci U S A. 2007;104(50):19983–8. doi: 10.1073/pnas.0706641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peng X, Guo W, Liu T, Wang X, Tu X, Xiong D, Chen S, Lai Y, Du H, Chen G, Liu G, Tang Y, Huang S, Zou X. Identification of miRs-143 and -145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. PLoS One. 2011;6(5):e20341. doi: 10.1371/journal.pone.0020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kong D, Li Y, Wang Z, Banerjee S, Ahmad A, Kim HR, Sarkar FH. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27(8):1712–21. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature cell biology. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 65.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids-the mix of hormones and biomarkers. Nature reviews. Clinical oncology. 2011;8(8):467–77. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goyal A, Delves GH, Chopra M, Lwaleed BA, Cooper AJ. Prostate cells exposed to lycopene in vitro liberate lycopene-enriched exosomes. BJU Int. 2006;98(4):907–11. doi: 10.1111/j.1464-410X.2006.06434.x. [DOI] [PubMed] [Google Scholar]
- 67.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 68.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 69.El-Hefnawy T, Raja S, Kelly L, Bigbee WL, Kirkwood JM, Luketich JD, Godfrey TE. Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin Chem. 2004;50(3):564–73. doi: 10.1373/clinchem.2003.028506. [DOI] [PubMed] [Google Scholar]
- 70.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Zen K, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 71.Liu CG, Calin GA, Volinia S, Croce CM. MicroRNA expression profiling using microarrays. Nat Protoc. 2008;3(4):563–78. doi: 10.1038/nprot.2008.14. [DOI] [PubMed] [Google Scholar]
- 72.Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA.: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–6. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 73.Leite KR, Tomiyama A, Reis ST, Sousa-Canavez JM, Sanudo A, Dall’oglio MF, Camara-Lopes LH, Srougi M. MicroRNA-100 Expression is Independently Related to Biochemical Recurrence of Prostate Cancer. J Urol. 2011;185(3):1118–22. doi: 10.1016/j.juro.2010.10.035. [DOI] [PubMed] [Google Scholar]
- 74.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67(13):6130–5. doi: 10.1.158/0008-5472.CAN-07-0533. 67/13/6130 (pii) [DOI] [PubMed] [Google Scholar]
- 76.Wang L, Tang H, Thayanithy V, Subramanian S, Oberg AL, Cunningham JM, Cerhan JR, Steer CJ, Thibodeau SN. Gene networks and microRNAs implicated in aggressive prostate cancer. Cancer Res. 2009;69(24):9490–7. doi: 10.1158/0008-5472.CAN-09-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martens-Uzunova ES, Jalava SE, Dits NF, van Leenders GJ, Moller S, Trapman J, Bangma CH, Litman T, Visakorpi T, Jenster G. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene. 2011;31(8):978–91. doi: 10.1038/onc.2011.304. [DOI] [PubMed] [Google Scholar]
- 78.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic. progress in validating and targeting microRNAs for cancer therapy. Nature reviews. Cancer. 2011;11(12):849–64. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gandellini P, Folini M, Longoni N, Pennati M, Binda M, Colecchia M, Salvioni R, Supino R, Moretti R, Limonta P, Valdagni R, Daidone MG, Zaffaroni N. miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 2009;69(6):2287–95. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 80.Li Y, VandenBoom TG, 2nd, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;696704–12(16) doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, Knyazev P, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7(16):2591–600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 82.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 17(2):193–9. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 83.Casanova-Salas I, Rubio-Briones J, Fernandez-Serra A, Lopez-Guerrero JA. miRNAs as biomarkers in prostate cancer. Clin Transl Oncol. 2012;14(11):803–11. doi: 10.1007/s12094-012-0877-0. [DOI] [PubMed] [Google Scholar]
- 84.Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang SL, Dai B, Zhu YP, Shen YJ, Shi GH, Ye DW. Serum miRNA-21. elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71(3):326–31. doi: 10.1002/pros.21246. [DOI] [PubMed] [Google Scholar]
- 85.Brase JC, Johannes M, Schlomm T, Falth M, Haese A, Steuber T, Beissbarth T, Kuner R, Sultmann H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128(3):608–16. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 86.Singh PK, Preus L, Hu Q, Yan L, Long MD, Morrison CD, Nesline M, Johnson CS, Koochekpour S, Kohli M, Liu S, Trump DL, Sucheston-Campbell LE, Campbell MJ. Serum microRNA expression patterns that predict early treatment failure in prostate cancer patients. Oncotarget. 2014;5(3):824–40. doi: 10.18632/oncotarget.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Theodore SC, Davis M, Zhao F, Wang H, Chen D, Rhim J, Dean-Colomb W, Turner T, Ji W, Zeng G, Grizzle W, Yates C. MicroRNA profiling of novel African American and Caucasian Prostate Cancer cell lines reveals a reciprocal regulatory relationship of miR-152 and DNA methyltranferase 1. Oncotarget. 2014;5(11):3512–25. doi: 10.18632/oncotarget.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lussier YA, Stadler WM, Chen JL. Advantages of genomic complexity: bioinformatics opportunities in microRNA cancer signatures. J Am Med Inform Assoc. 2012;19(2):156–60. doi: 10.1136/amiajnl-2011-000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tomlins SA, Day JR, Lonigro RJ, Hovelson DH, Siddiqui J, Kunju LP, Dunn RL, Meyer S, Hodge P, Groskopf J, Wei JT, Chinnaiyan AM. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur Urol. 2015 doi: 10.1016/j.eururo.2015.04.039. pii: S0302-2838(15)00397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Young A, Palanisamy N, Siddiqui J, Wood DP, Wei JT, Chinnaiyan AM, Kunju LP, Tomlins SA. Correlation of urine TMPRSS2:ERG and PCA3 to ERG+ and total prostate cancer burden. Am J Clin Pathol. 2012;138(5):685–96. doi: 10.1309/AJCPU7PPWUPYG8OH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M, Gong Y, Cheng H, Laxman B, Vellaichamy A, Shankar S, Li Y, Dhanasekaran SM, Morey R, Barrette T, Lonigro RJ, Tomlins SA, Varambally S, Qin ZS, Chinnaiyan AM. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17(5):443–54. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R, Lonigro RJ, Tsodikov A, Wei JT, Tomlins SA, Chinnaiyan AM. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68(3):645–9. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Demichelis F, Fall K, Perner S, Andren O, Schmidt F, Setlur SR, Hoshida Y, Mosquera JM, Pawitan Y, Lee C, Adami HO, Mucci LA, Kantoff PW, Andersson SO, Chinnaiyan AM, Johansson JE, Rubin MA. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26(31):4596–9. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 94.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 95.Vaarala MH, Porvari K, Kyllonen A, Lukkarinen O, Vihko P. The TMPRSS2 gene encoding transmembrane serine protease is overexpressed in a majority of prostate cancer patients: detection of mutated TMPRSS2 form in a case of aggressive disease. Int J Cancer. 2001;94(5):705–10. doi: 10.1002/ijc.1526. [DOI] [PubMed] [Google Scholar]
- 96.Lubieniecka JM, Cheteri MK, Stanford JL, Ostrander EA. Met160Val polymorphism in the TRMPSS2 gene and risk of prostate cancer in a population-based case-control study. Prostate. 2004;59(4):357–9. doi: 10.1002/pros.20005. [DOI] [PubMed] [Google Scholar]
- 97.Farrell J, Young D, Chen Y, Cullen J, Rosner IL, Kagan J, Srivastava S, Mc LD, Sesterhenn IA, Srivastava S, Petrovics G. Predominance of ERG-negative high-grade prostate cancers in African American men. Mol Clin Oncol. 2014;2(6):982–986. doi: 10.3892/mco.2014.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.St John J, Powell K, Conley-Lacomb MK, Chinni SR. TMPRSS2-ERG Fusion Gene Expression in Prostate Tumor Cells and Its Clinical and Biological Significance in Prostate Cancer Progression. J Cancer Sci Ther. 2012;4(4):94–101. doi: 10.4172/1948-5956.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma W, Diep K, Fritsche HA, Shore N, Albitar M. Diagnostic and prognostic scoring system for prostate cancer using urine and plasma biomarkers. Genet Test Mol Biomarkers. 2014;18(3):156–63. doi: 10.1089/gtmb.2013.0424. [DOI] [PubMed] [Google Scholar]
- 100.Dijkstra S, Birker IL, Smit FP, Leyten GH, de Reijke TM, van Oort IM, Mulders PF, Jannink SA, Schalken JA. Prostate cancer biomarker profiles in urinary sediments and exosomes. J Urol. 2014;191(4):1132–8. doi: 10.1016/j.juro.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 101.Lin DW, Newcomb LF, Brown EC, Brooks JD, Carroll PR, Feng Z, Gleave ME, Lance RS, Sanda MG, Thompson IM, Wei JT, Nelson PS I. Canary Prostate Active Surveillance Study. Urinary TMPRSS2:ERG and PCA3 in an active surveillance cohort: results from a baseline analysis in the Canary Prostate Active Surveillance Study. Clin Cancer Res. 2013;19(9):2442–50. doi: 10.1158/1078-0432.CCR-12-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Park K, Dalton JT, Narayanan R, Barbieri CE, Hancock ML, Bostwick DG, Steiner MS, Rubin MA. TMPRSS2:ERG gene fusion predicts subsequent detection of prostate cancer in patients with high-grade prostatic intraepithelial neoplasia. J Clin Oncol. 2014;32(3):206–11. doi: 10.1200/JCO.2013.49.8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Casey OM, Fang L, Hynes PG, Abou-Kheir WG, Martin PL, Tillman HS, Petrovics G, Awwad HO, Ward Y, Lake R, Zhang L, Kelly K. TMPRSS2- driven ERG expression in vivo increases self-renewal and maintains expression in a castration resistant subpopulation. PLoS One. 2012;7(7):e41668. doi: 10.1371/journal.pone.0041668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Williamson SR, Zhang S, Yao JL, Huang J, Lopez-Beltran A, Shen S, Osunkoya AO, MacLennan GT, Montironi R, Cheng L. ERG-TMPRSS2 rearrangement is shared by concurrent prostatic adenocarcinoma and prostatic small cell carcinoma and absent in small cell carcinoma of the urinary bladder: evidence supporting monoclonal origin. Mod Pathol. 2011;24(8):1120–7. doi: 10.1038/modpathol.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li Y, Kong D, Wang Z, Ahmad A, Bao B, Padhye S, Sarkar FH. Inactivation of AR/TMPRSS2-ERG/Wnt signaling networks attenuates the aggressive behavior of prostate cancer cells. Cancer Prev Res (Phila) 2011;4(9):1495–506. doi: 10.1158/1940-6207.CAPR-11-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brase JC, Johannes M, Mannsperger H, Falth M, Metzger J, Kacprzyk LA, Andrasiuk T, Gade S, Meister M, Sirma H, Sauter G, Simon R, Schlomm T, Beissbarth T, Korf U, Kuner R, Sultmann H. TMPRSS2-ERG -specific transcriptional modulation is associated with prostate cancer biomarkers and TGF-beta signaling. BMC Cancer. 2011;11:507. doi: 10.1186/1471-2407-11-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rubio-Briones J, Fernandez-Serra A, Calatrava A, Garcia-Casado Z, Rubio L, Bonillo MA, Iborra I, Solsona E, Lopez-Guerrero JA. Clinical implications of TMPRSS2-ERG gene fusion expression in patients with prostate cancer treated with radical prostatectomy. J Urol. 2010;183(5):2054–61. doi: 10.1016/j.juro.2009.12.096. [DOI] [PubMed] [Google Scholar]
- 108.Saramaki OR, Harjula AE, Martikainen PM, Vessella RL, Tammela TL, Visakorpi T. TMPRSS2:ERG fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin Cancer Res. 2008;14(11):3395–400. doi: 10.1158/1078-0432.CCR-07-2051. [DOI] [PubMed] [Google Scholar]
- 109.Magi-Galluzzi C, Tsusuki T, Elson P, Simmerman K, LaFargue C, Esgueva R, Klein E, Rubin MA, Zhou M. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate. 2011;71(5):489–97. doi: 10.1002/pros.21265. [DOI] [PubMed] [Google Scholar]
- 110.Taris M, Irani J, Blanchet P, Multigner L, Cathelineau X, Fromont G. ERG expression in prostate cancer: the prognostic paradox. Prostate. 2014;74(15):1481–7. doi: 10.1002/pros.22863. [DOI] [PubMed] [Google Scholar]
- 111.Rawal S, Young D, Williams M, Colombo M, Krishnappa R, Petrovics G, McLeod DG, Srivastava S, Sesterhenn IA. Low Frequency of the ERG Oncogene Alterations in Prostate Cancer Patients from India. J Cancer. 2013;4(6):468–72. doi: 10.7150/jca.6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Magi-Galluzzi C, Tsusuki T, Elson P, Simmerman K, LaFargue C, Esgueva R, Klein E, Rubin MA, Zhou M. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate. 2011;71(5):489–97. doi: 10.1002/pros.21265. [DOI] [PubMed] [Google Scholar]
- 113.Giri VN, Ruth K, Hughes L, Uzzo RG, Chen DY, Boorjian SA, Viterbo R, Rebbeck TR. Racial differences in prediction of time to prostate cancer diagnosis in a prospective screening cohort of high-risk men: effect of TMPRSS2 Met160Val. BJU Int. 2011;107(3):466–70. doi: 10.1111/j.1464-410X.2010.09522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Braun M, Goltz D, Shaikhibrahim Z, Vogel W, Bohm D, Scheble V, Sotlar K, Fend F, Tan SH, Dobi A, Kristiansen G, Wernert N, Perner S. ERG protein expression and genomic rearrangement status in primary and metastatic prostate cancer--a comparative study of two monoclonal antibodies. Prostate Cancer Prostatic Dis. 2012;15(2):165–9. doi: 10.1038/pcan.2011.67. [DOI] [PubMed] [Google Scholar]
- 115.Rosen P, Sesterhenn IA, Brassell SA, McLeod DG, Srivastava S, Dobi A. Clinical potential of the ERG oncoprotein in prostate cancer. Nat Rev Urol. 2012;9(3):131–7. doi: 10.1038/nrurol.2012.10. [DOI] [PubMed] [Google Scholar]
- 116.Falzarano SM, Zhou M, Carver P, Tsuzuki T, Simmerman K, He H, Magi-Galluzzi C. ERG gene rearrangement status in prostate cancer detected by immunohistochemistry. Virchows Arch. 2011;459(4):441–7. doi: 10.1007/s00428-011-1128-4. [DOI] [PubMed] [Google Scholar]
- 117.Farrell J, Petrovics G, McLeod DG, Srivastava S. Genetic and molecular differences in prostate carcinogenesis between African American and Caucasian American men. Int J Mol Sci. 2013;14(8):15510–31. doi: 10.3390/ijms140815510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rosen P, Pfister D, Young D, Petrovics G, Chen Y, Cullen J, Bohm D, Perner S, Dobi A, McLeod DG, Sesterhenn IA, Srivastava S. Differences in frequency of ERG oncoprotein expression between index tumors of Caucasian and African American patients with prostate cancer. Urology. 2012;80(4):749–53. doi: 10.1016/j.urology.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rice KR, Chen Y, Ali A, Whitman EJ, Blase A, Ibrahim M, Elsamanoudi S, Brassell S, Furusato B, Stingle N, Sesterhenn IA, Petrovics G, Miick S, Rittenhouse H, Groskopf J, McLeod DG, Srivastava S. Evaluation of the ETS-related gene mRNA in urine for the detection of prostate cancer. Clin Cancer Res. 2010;16(5):1572–6. doi: 10.1158/1078-0432.CCR-09-2191. [DOI] [PubMed] [Google Scholar]
- 120.Yamoah K, Johnson MH, Choeurng V, Faisal FA, Yousefi K, Haddad Z, Ross AE, Alshalafa M, Den R, Lal P, Feldman M, Dicker AP, Klein EA, Davicioni E, Rebbeck TR, Schaeffer EM. Novel Biomarker Signature That May Predict Aggressive Disease in African American Men With Prostate Cancer. J Clin Oncol. 2015;33(25):2789–96. doi: 10.1200/JCO.2014.59.8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Martin DN, Starks AM, Ambs S. Biological determinants of health disparities in prostate cancer. Current opinion in oncology. 2013;25(3):235–41. doi: 10.1097/cco.0b013e32835eb5d1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kwabi-Addo B, Wang S, Chung W, Jelinek J, Patierno SR, Wang BD, Andrawis R, Lee NH, Apprey V, Issa JP, Ittmann M. Identification of differentially methylated genes in normal prostate tissues from African American and Caucasian men. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16(14):3539–47. doi: 10.1158/1078-0432.CCR-09-3342. [DOI] [PubMed] [Google Scholar]
- 123.Enokida H, Shiina H, Urakami S, Igawa M, Ogishima T, Pookot D, Li LC, Tabatabai ZL, Kawahara M, Nakagawa M, Kane CJ, Carroll PR, Dahiya R. Ethnic group-related differences in CpG hypermethylation of the GSTP1 gene promoter among African-American, Caucasian and Asian patients with prostate cancer. International journal of cancer. Journal international du cancer. 2005;116(2):174–81. doi: 10.1002/ijc.21017. [DOI] [PubMed] [Google Scholar]
- 124.Woodson K, Hayes R, Wideroff L, Villaruz L, Tangrea J. Hypermethylation of GSTP1, CD44, and E-cadherin genes in prostate cancer among US Blacks and Whites. Prostate. 2003;55(3):199–205. doi: 10.1002/pros.10236. [DOI] [PubMed] [Google Scholar]
- 125.Kwabi-Addo B, Wang S, Chung W, Jelinek J, Patierno SR, Wang BD, Andrawis R, Lee NH, Apprey V, Issa JP, Ittmann M. Identification of differentially methylated genes in normal prostate tissues from African American and Caucasian men. Clin Cancer Res. 2010;16(14):3539–47. doi: 10.1158/1078-0432.CCR-09-3342. [DOI] [PubMed] [Google Scholar]
- 126.Kobayashi Y, Absher DM, Gulzar ZG, Young SR, McKenney JK, Peehl DM, Brooks JD, Myers RM, Sherlock G. DNA methylation profiling reveals novel biomarkers and important roles for DNA methyltransferases in prostate cancer. Genome Res. 2011;21(7):1017–27. doi: 10.1101/gr.119487.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. 2012;31(13):1609–22. doi: 10.1038/onc.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yaqinuddin A, Qureshi SA, Qazi R, Farooq S, Abbas F. DNMT1 silencing affects locus specific DNA methylation and increases prostate cancer derived PC3 cell invasiveness. J Urol. 2009;182(2):756–61. doi: 10.1016/j.juro.2009.03.082. [DOI] [PubMed] [Google Scholar]
- 129.Ho VT, Vanneman M, Kim H, Sasada T, Kang YJ, Pasek M, Cutler C, Koreth J, Alyea E, Sarantopoulos S, Antin JH, Ritz J, Canning C, Kutok J, Mihm MC, Dranoff G, Soiffer R. Biologic activity of irradiated, autologous, GM-CSF-secreting leukemia cell vaccines early after allogeneic stem cell transplantation. Proc Natl Acad Sci U S A. 2009;106(37):15825–30. doi: 10.1073/pnas.0908358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Borno ST, Fischer A, Kerick M, Falth M, Laible M, Brase JC, Kuner R, Dahl A, Grimm C, Sayanjali B, Isau M, Rohr C, Wunderlich A, Timmermann B, Claus R, Plass C, Graefen M, Simon R, Demichelis F, Rubin MA, Sauter G, Schlomm T, Sultmann H, Lehrach H, Schweiger MR. Genome-wide DNA methylation events in TMPRSS2-ERG fusion-negative prostate cancers implicate an EZH2-dependent mechanism with miR-26a hypermethylation. Cancer Discov. 2012;2(11):1024–35. doi: 10.1158/2159-8290.CD-12-0041. [DOI] [PubMed] [Google Scholar]
- 131.Zhan C, Yan L, Wang L, Jiang W, Zhang Y, Xi J, Chen L, Jin Y, Qiao Y, Shi Y, Wang Q. Identification of reference miRNAs in human tumors by TCGA miRNA-seq data. Biochem Biophys Res Commun. 2014;453(3):375–8. doi: 10.1016/j.bbrc.2014.09.086. [DOI] [PubMed] [Google Scholar]
- 132.Cline MS, Craft B, Swatloski T, Goldman M, Ma S, Haussler D, Zhu J. Exploring TCGA Pan-Cancer data at the UCSC Cancer Genomics Browser. Sci Rep. 2013;3:2652. doi: 10.1038/srep02652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang RS, Gamazon ER, Ziliak D, Wen Y, Im HK, Zhang W, Wing C, Duan S, Bleibel WK, Cox NJ, Dolan ME. Population differences in microRNA expression and biological implications. RNA biology. 2011;8(4):692–701. doi: 10.4161/rna.8.4.16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy. 2012;8(2):165–76. doi: 10.4161/auto.8.2.18351. [DOI] [PubMed] [Google Scholar]
- 135.Li LJ, Huang Q, Zhang N, Wang GB, Liu YH. miR-376b-5p regulates angiogenesis in cerebral ischemia. Mol Med Rep. 2014;10(1):527–35. doi: 10.3892/mmr.2014.2172. [DOI] [PubMed] [Google Scholar]
- 136.Yang X, Bemis L, Su LJ, Gao D, Flaig TW. miR-125b Regulation of Androgen Receptor Signaling Via Modulation of the Receptor Complex Co-Repressor NCOR2. BioResearch open access. 2012;1(2):55–62. doi: 10.1089/biores.2012.9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang H, Liu W, Black S, Turner O, Daniel JM, Dean-Colomb W, He QP, Davis M, Yates C. Kaiso, a transcriptional repressor, promotes cell migration and invasion of prostate cancer cells through regulation of miR-31 expression. Oncotarget. 2015 doi: 10.18632/oncotarget.6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jones J, Wang H, Zhou J, Hardy S, Turner T, Austin D, He Q, Wells A, Grizzle WE, Yates C. Nuclear Kaiso indicates aggressive prostate cancers and promotes migration and invasiveness of prostate cancer cells. Am J Pathol. 2012;181(5):1836–46. doi: 10.1016/j.ajpath.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Formosa A, Lena AM, Markert EK, Cortelli S, Miano R, Mauriello A, Croce N, Vandesompele J, Mestdagh P, Finazzi-Agro E, Levine AJ, Melino G, Bernardini S, Candi E. DNA methylation silences miR-132 in prostate cancer. Oncogene. 2013;32(1):127–34. doi: 10.1038/onc.2012.14. [DOI] [PubMed] [Google Scholar]
- 140.Huan T, Rong J, Liu C, Zhang X, Tanriverdi K, Joehanes R, Chen BH, Murabito JM, Yao C, Courchesne P, Munson PJ, O’Donnell CJ, Cox N, Johnson AD, Larson MG, Levy D, Freedman JE. Genome-wide identification of microRNA expression quantitative trait loci. Nat Commun. 2015;6:6601. doi: 10.1038/ncomms7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–7. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]