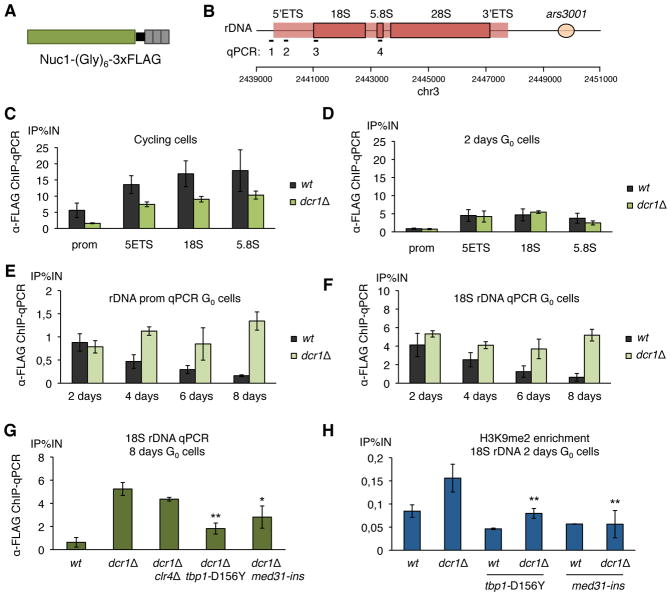
Figure 4. dcr1Δ mutants are defective in RNA polymerase I release in quiescence maintenance.
(A) The main subunit of RNA polymerase I Nuc1RPA190 was tagged with a (Gly)6 linker and 3xFLAG. (B) Probe location for ChIP-qPCRs: 1: rDNA promoter, 2: 5′ETS, 3: 18S, 4: 3′ETS. (C) RNA polymerase I enrichment at the rDNA was assayed by ChIP-qPCR using anti-FLAG antibody in the nuc1-(Gly)6-FLAG background, showing an higher occupancy in wild-type than dcr1Δ mutants in cycling cells, due to the reduced growth rate of dcr1Δ, and (D) a similar occupancy in early G0 cells (n=2 biological replicates). (E, F) RNA polymerase I enrichment at 18S rDNA over time spent in G0 (2 to 8 days) shows a reduction in wild-type cells, but dcr1Δ cells fail to release RNA pol I (n≥2 biological replicates). (G) The class (iii) suppressors, dcr1Δ tbp1-D156Y and dcr1Δmed31-ins, but not the class (ii) suppressor dcr1Δclr4Δ, significantly decrease RNA polymerase I occupancy at rDNA in 8 days G0 cells. (n=2 biological replicates, * p<0.05, ** p<0.01, t-test) (H) The accumulation of H3K9me2 at rDNA repeats, assayed by ChIP-qPCR, is significantly reduced in class (iii) suppressors (n≥3 biological replicates, ** p<0.01, t-test).