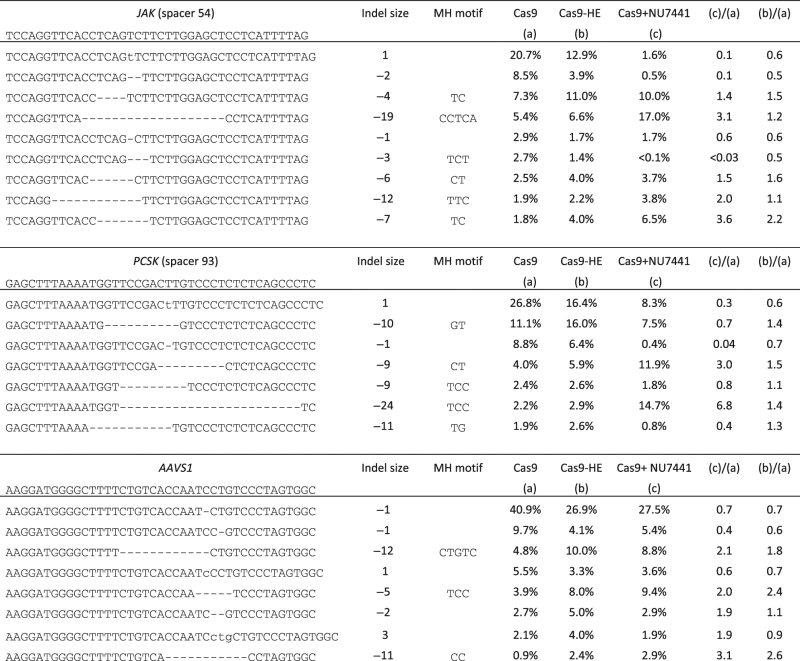
Table 1.
Indel mutation patterns induced by Cas9, Cas9–HE, and Cas9+NU7441
|
Indel mutation patterns induced after transfection of nucleases and guide RNA expression vectors were determined by sequencing of PCR amplicons of the targeted region. The wild-type target sequence is indicated on top of mutant sequences. When indicated, cells were treated with 10 μM DNA-PK inhibitor NU7441
The indels shown are indels that represented more than 2% of mutant reads obtained with Cas9 or Cas9–HE. If present, microhomologies (MH) of two or more nucleotides flanking the deletion are indicated
Spacer 54 and Spacer 93 are from guide RNAs previously analyzed by van Overbeek et al.21
For spacer 54, mutant reads were 35.7% (of the total 47,199 reads), 29.8% (of the total 48,265 reads), and 6.5% (of the total 116,354 reads) for Cas9, Cas9–HE, and Cas9+NU7441, respectively. For spacer 93, mutant reads were 31.3% (of the total 45,398 reads), 24.2% (of the total 55,573 reads), and 4.1% (of the total 36,979 reads) for Cas9, Cas9–HE, and Cas9+NU7441, respectively. For T2 guide RNA, mutant reads were 39% (of the total 68,852 reads), 16.8% (of the total 67,815 reads), and 31.8% (of the total 69,696 reads) for Cas9, Cas9–HE, and Cas9+NU7441, respectively
