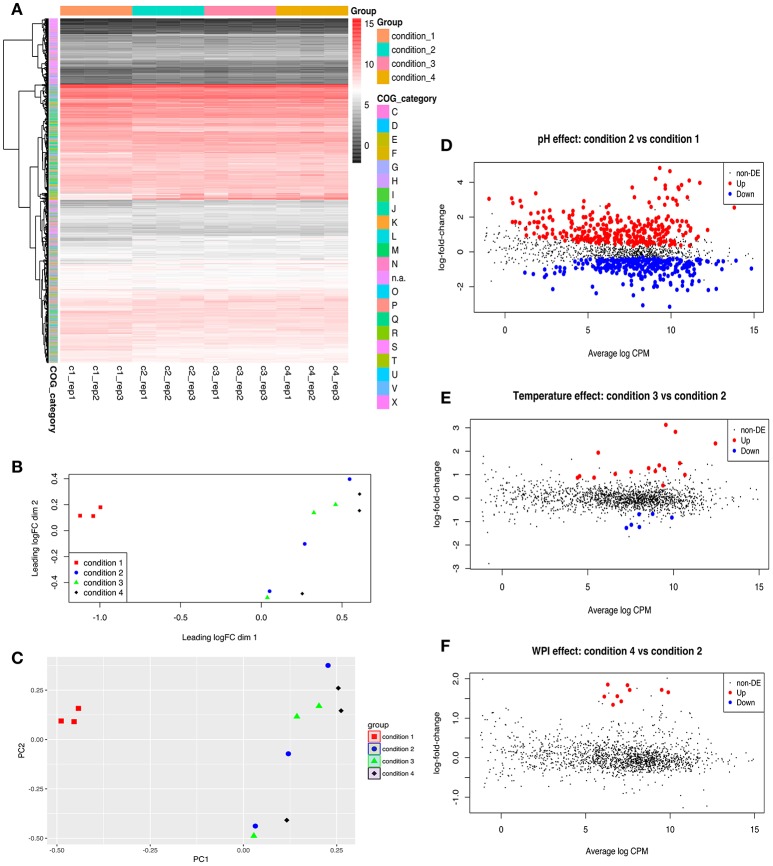
Figure 1.
Genome-wide comparative transcriptome analysis of ST1275 under different pH, temperature and milk base. (A) Heatmap of gene expression level for samples collected from 4 conditions; genes are retained if they are expressed at a count-per-million (CPM) above 0.5 in at least two samples; expression values are normalized log2CPM by using edgeR package. (B) visualization of expression profiles (log2CPM) of different samples by multi-dimensional scaling (MDS) plot in edgeR package. (C) visualization of expression profiles (log2CPM) of different samples by principal component analysis (PCA) plot in ggfortify package. (D) Detection of DEGs for samples from condition 2 compared to samples from condition 1. (E) Detection of DEGs for samples from condition 3 compared to samples from condition 2. (F) Detection of DEGs for samples from condition 4 compared to samples from condition 2. Genes with a fold-change significantly above 1.0 at an FDR cut-off of 5% is considered as DEGs. COG denotation: C, energy production and conversion; D, cell cycle control, cell division, chromosome partitioning; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; M, cell wall/membrane/envelope biogenesis; N, cell motility; n.a., not assigned; O, post-translational modification, protein turnover, chaperones; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown; T, signal transduction mechanisms; U, intracellular trafficking, secretion, and vesicular transport; V, defense mechanisms; X, mobilome: prophages, transposons.