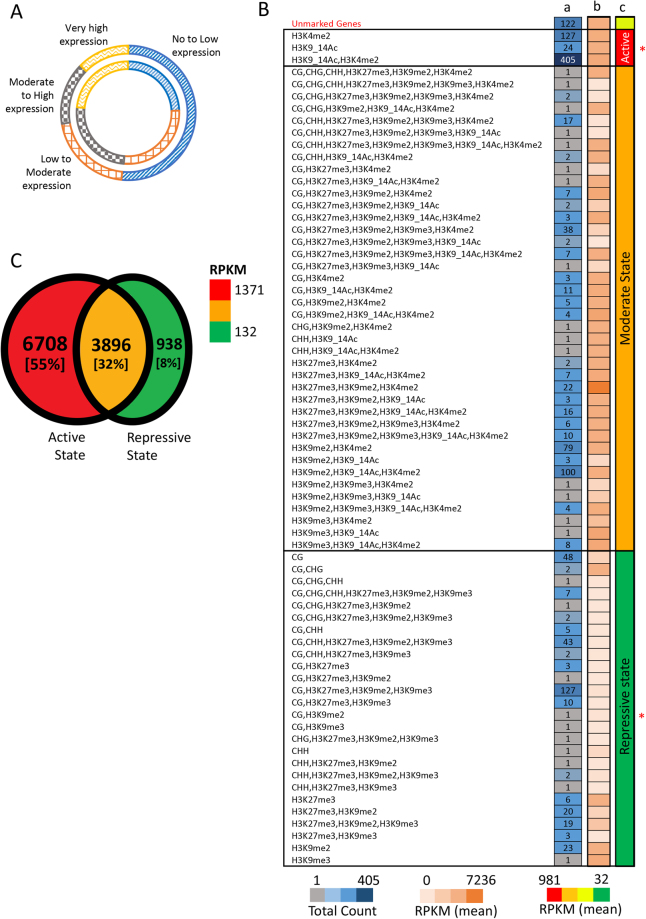
Figure 3.
Dissecting the functional characteristics of Phatr3 proteome. (A) Comparison of the expression profile of the proportion of new genes (outer circle) with that of the proportion of unchanged gene models (inner circle). The expression profiling of all the Phatr3 genes are done based on quartile method, where first and last quartile reflects genes with no/low expression and very high expression, respectively. (B) and (C) summarize various epigenetic modification and regulatory effects on Phatr3 gene models and only new gene models in Phatr3. (B) Specifies the regulatory effects of the co-localization of different modifications/marks on the novel genes. The number of genes corresponding to each combination of co-localizing marks is provided in column a. The average expression of these genes is represented as a heat map in column b. Compared to the unmarked genes the regulatory effect of the co-localization of different epigenetic modifications is seen to maintain three chromatin states, represented with column c with few exceptions with expression that can be higher or lower than expected. This cases are likely due to the presence of additional active or repressive marks that were not studied. *Indicates where the average expression (normalized DESeq counts) is significantly different (two sample T-test with unequal variance, P-value < 0.002), compared to the expression of unmarked genes. Out of 1,489 genes, 1,388 are considered in the analysis, as expression for the other 101 genes was not calculable without errors. (C) Different chromatin states maintained genome wide, based on the association of protein coding genes with repressive, active or both repressive and active chromatin modifiers (Repressive modifications: CG, CHG, CHH, H3K27me3, H3K9me2, H3K9me3; Active modifications: H3K4me2, H3K9_14Ac). Numbers and percentages (in square brackets) in the Venn diagram reflects the absolute number of genes and the relative percentage of the total Phatr3 genes.