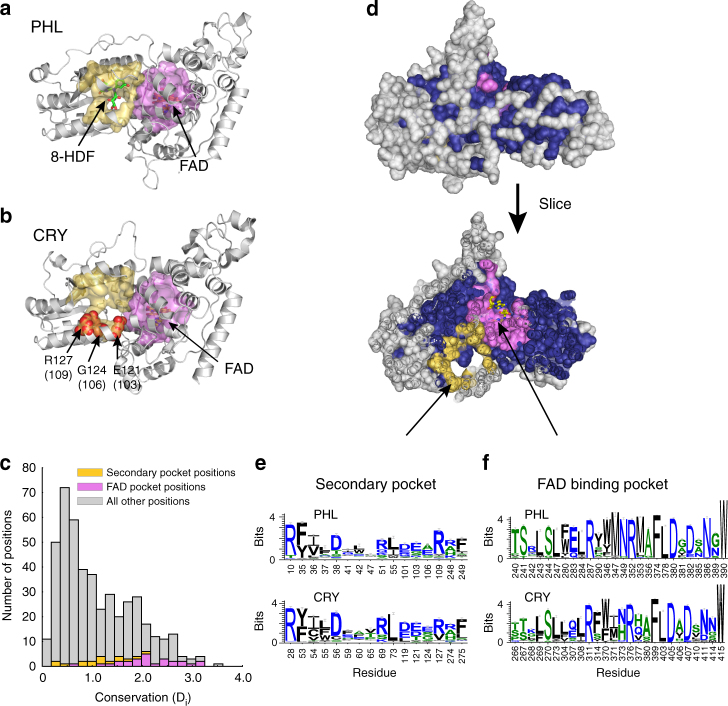
Fig. 1.
Identification of an allosteric secondary pocket in the CRYs by comparison to the PHLs. a The PHL domain is shown in gray cartoon (PDB: 1TEZ). The secondary pocket (yellow surface) and primary FAD-binding pocket (pink surface) were defined as all residues with at least one atom within 4 Å of 8-hydroxy-7,8-didemethyl-5-deazariboflavin (8-HDF) (green sticks) or FAD (yellow sticks) respectively. b The FAD-binding and secondary pockets mapped to the CRY2 structure (PDB: 4I6G). Three positions within the secondary pocket were previously identified in a functional screen (highlighted as red spheres). c Distribution of conservation values for all positions in the CRY/photolyase family (CPF) alignment. Conservation is computed as the Kullback–Leibler relative entropy (Di), which measures the divergence of amino acid frequencies at a particular position from the distribution expected by random chance. Values near zero indicate that the observed frequencies at a site are close to random expectation, while values above three reflect positions approaching near-total conservation (84% identity or more). The majority of positions are weakly conserved; the secondary pocket positions (yellow) are moderately conserved, and the FAD-binding pocket positions (pink) are more conserved. d Mapping of the sector (dark blue spheres) to the mCRY2 structure. Non-sector positions are shown in gray spheres, and the primary and secondary pockets are again indicated in pink and yellow spheres respectively. e–f Sequence logo plots describing the sequence variation in the FAD-binding pocket and secondary pocket. Residue numbers correspond to the structures shown in a and b. The sequence variation is computed for sequences annotated as either CRYs or PHLs. We observe a similar degree of conservation within the CRYs and PHLs for both pockets