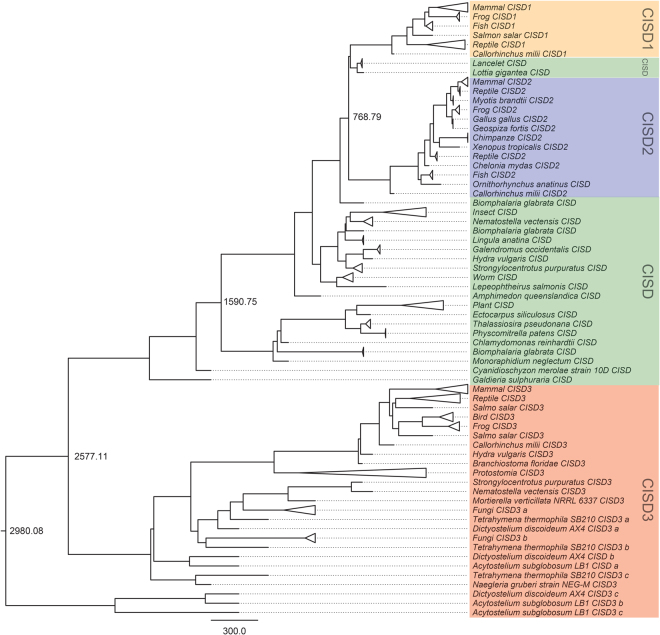
Figure 5.
Molecular clock analysis of CDGSH evolution in eukaryotes. A time tree of divergence among eukaryotic CISD proteins constructed using the BEAST (Bayesian Evolutionary Analysis Sampling Trees) software. Eukaryotic sequences were obtained as described in the Methods section. A total of 150 sequences were subjected to Bayesian analysis using two model combinations (Supplementary Table S3. Supplementary Material) - a constant (shown here and Supplementary Fig. S9) and an exponential (Supplementary Fig. S10) population size model with a relaxed uncorrelated log-normal clock. The divergence times for Class I/II and Class III, Class I and Class II, and the putative ancestor of eukaryotic CISD proteins are shown. An expanded version of the two trees (constant and exponential) with complete protein annotations and estimated node ages is shown in Supplementary Figs S9 and S10.