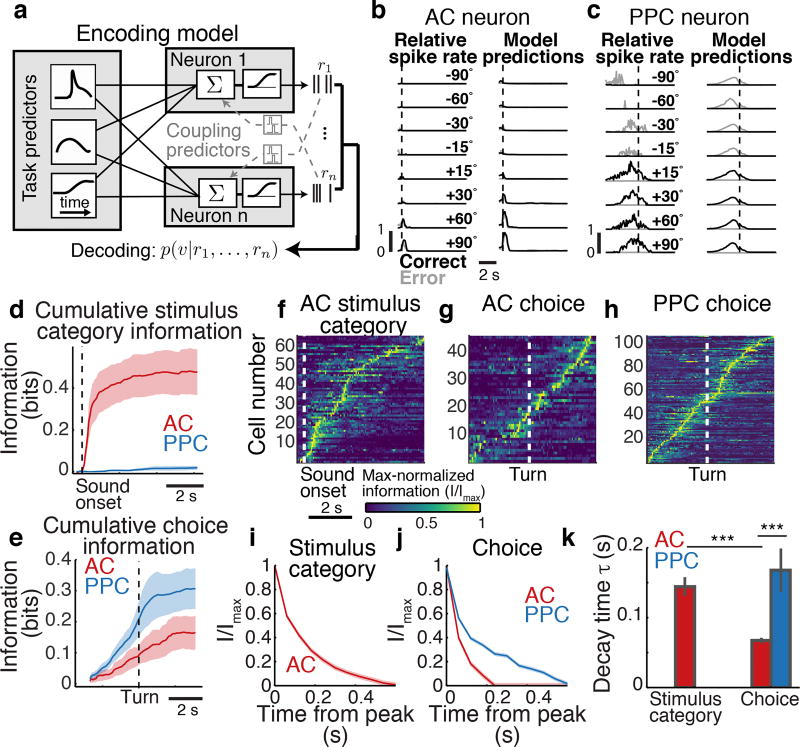
Figure 2. Encoding and decoding stimulus and choice information in AC and PPC.
a, The encoding model (GLM) was fit to each neuron’s activity using task predictors (“uncoupled model”) or task predictors and activity of other neurons (coupling predictors, “coupled model”). To decode stimuli or choices (indicated as ν), the posterior probability of each stimulus or choice was computed, based on the current time point only (“instantaneous decoder”), or all previous time points in the trial (“cumulative decoder”). The decoder could use either all neurons (population decoder) or individual neurons (single-neuron decoder). b–c, Example trial-averaged responses (left column) and model predictions (right column) in correct (black) and error (gray) trials. d–e, Cumulative stimulus category and choice information decoded in AC (red) and PPC (blue) populations. Shading indicates mean ± sem across datasets. n = 7 datasets for AC and PPC. f–h, Max-normalized instantaneous information about stimulus category and choice for AC and PPC cells with at least 0.06 bits of information at any point in the trial (fraction of all cells: AC: stimulus, 20.2 ± 5.4%; choice, 15.9 ± 3.2%; PPC: stimulus, 3.1 ± 1.3%; choice, 19.3 ± 4.6%). Neurons were sorted by the times of their maximum information. i–j, Information about stimulus category and choice averaged across all AC (red) and PPC (blue) cells, calculated as a two-sided decay (forward and backward in time) around the information peak of each cell. Shading indicates mean ± sem across cells with at least 0.06 bits of information. k, Decay time constant of single-cell information from exponential fits to information time courses in (i–j). Error bars: 95% confidence intervals (Methods). *** indicates p < 0.001, z-test.