Abstract
Objective
Rapid advances of high-throughput technologies and wide adoption of electronic health records (EHRs) have led to fast accumulation of -omic and EHR data. These voluminous complex data contain abundant information for precision medicine, and big data analytics can extract such knowledge to improve the quality of health care.
Methods
In this article, we present -omic and EHR data characteristics, associated challenges, and data analytics including data pre-processing, mining, and modeling.
Results
To demonstrate how big data analytics enables precision medicine, we provide two case studies, including identifying disease biomarkers from multi-omic data and incorporating -omic information into EHR.
Conclusion
Big data analytics is able to address –omic and EHR data challenges for paradigm shift towards precision medicine.
Significance
Big data analytics makes sense of –omic and EHR data to improve healthcare outcome. It has long lasting societal impact.
Index Terms: Precision medicine, big data analytics, -omic data, electronic health records, bioinformatics, health informatics
I. Introduction
TO achieve the best care for patients, many models have been proposed over the years to improve the healthcare system. The goal of the early “personalized medicine” model is to customize healthcare delivery for each individual and to maximize the effectiveness of each patient’s treatment [1]. In 2009, Hood et al. propose the “personalized, predictive, preventive, and participatory medicine” (a.k.a. P4 medicine) model that aims to transform current reactive care to future proactive medicine, and ultimately to reduce healthcare expenditure and improve patients’ health outcome [2]. Recently, the new “precision medicine” model is proposed to precisely classify patients into subgroups sharing a common biological basis of diseases for more effective treatment and improved care outcome [3, 4]. Precision medicine requires data utility ranging from collection and management (i.e. data storage, sharing, and privacy) to analytics (i.e. data mining, integration, and visualization) [5]. Because of rapid advances in biotechnologies, highly complex biomedical data are becoming available in huge volumes [6]. To make sense of these heterogeneous data, big data analytics, including data quality control, analysis, modeling, interpretation, and validation, is needed to cover application areas such as bioinformatics [7–9], health informatics [10–12], imaging informatics [13, 14], and sensor informatics [15, 16].
As presented in 2015 US Precision Medicine Initiative [17], incorporating -omic data and knowledge into electronic health record (EHR) (Fig. 1) is viewed as a necessary step for delivering precision medicine [3, 5, 17, 18]. Thus, this article reviews big -omic and EHR data analytics for precision medicine with key terms summarized in Tables I and II. Section II presents -omic and EHR data characteristics, challenges, and big data analytics; Section III uses two case studies to illustrate the impact of big data analytics in precision medicine; Section IV enumerates several well-known biomedical big data initiatives; Section V discusses current opportunities in big data analytics for precision medicine; and finally, Section VI concludes this article.
Fig. 1.
The key types of biomedical big data for precision medicine.
TABLE I.
Biomedical Big Data Keywords
| Topics | Keywords |
|---|---|
| -Omic Data | Genomics, transcriptomics, epigenomics, proteomics, metabolomics, etc. |
| EHR Data | Big data in EHR, next-generation EHR, clinical data management, medical coding systems, etc. |
| Data Challenges | Biomedical big data challenges, -omic data challenges, EHR data challenges, etc. |
| -Omic Data Analytics | NGS sequence mapping, NGS variant detection, RNA-seq computation, ChIP-seq computation, MS pre-processing, NGS biomarker identification, NGS differential analysis, -omic network analysis, -omic dynamic modeling, etc. |
| EHR Data Analytics | Temporal medical data mining, irregular time series analysis in EHR, clinical decision support, unsupervised/ supervised learning in EHR, waveform analysis in EHR, etc. |
| Big Data Analytics Enablers |
Big data harmonization, big data platform, big data framework |
TABLE II.
-Omic and EHR Data Concept Glossary
| Term | Definition | Ref. | |
|---|---|---|---|
| Genome | “An organism’s complete set of DNA” | [20] | |
| Transcriptome | “A collection of all the gene readouts present in an organism’s cell” |
[20] | |
| Epigenome | “A multitude of chemical compounds that can tell the genome what to do” |
[20] | |
| Proteome | “An entire set of proteins encoded by the genome” | [21] | |
| Metabolome | “A comprehensive catalogue of metabolites in an organism’s cell” |
[21] | |
| -Omics | “The study of the -ome” | [21] | |
| Single Nucleotide Polymorphism |
“A variation at a single position in a DNA sequence among individuals” |
[22] | |
| Frameshift Mutation (Indels) |
“A genetic mutation caused by a deletion or insertion in a DNA sequence that shifts the way the sequence is read” |
[22] | |
| Copy Number Variation |
“The number of copies of a particular genetic sequence differs between individuals” |
[22] | |
| Structural Variation |
“Genomic alterations that involve segments of DNA that are larger than 1 kb, and can be microscopic or submicroscopic” |
[23] | |
| Fusion Gene | “A new gene formed by the breakage and re-joining of two different genes” |
[24] | |
| Spanning Read Pair |
“paired reads that harbor a fusion boundary in the insert sequence” |
[25] | |
| Split Read | “A read that harbors a fusion boundary in the read itself ” | [25] | |
| Alternative Splicing |
A process that includes or excludes certain exons when forming mature mRNAs |
[26] | |
| Protein-DNA Binding Site |
A segment of DNA sequences where targeted proteins may bind |
[27] | |
| Histone Modification |
“A covalent post-translational modification to histone proteins that can impact gene expression” |
[27] | |
| DNA Methylation | “The addition of methyl (CH3) group to DNA that modifies the function of the genes” |
[27] | |
| Antecedent (Ant.) in ARL |
A set of conditions which the outcome variable depends on | [28] | |
| Consequent (Cons.) in ARL |
A set of conditions serving as the outcome variable | [28] | |
| Confidence in ARL |
[28] | ||
| Fast Healthcare Interoperability Resources (FHIR) |
FHIR uses standardized “Resources” (i.e., predefined data formats and elements) to exchange EHR data. |
[29] |
II. Big Data For Precision Medicine
The invention of high-throughput -omic assays such as next-generation sequencing (NGS) and mass spectrometry (MS) has led to fast accumulation of -omic data. Likewise, the wide adoption of EHR for the entire population provides a foundation for studying healthcare efficiency and safety [19]. -Omic data analytics often aims at finding biomarkers by cleaning up raw data generated by NGS or MS, extracting molecular profiles, identifying statistically significant molecules, constructing models describing molecular interactions or temporal system behavior, and validating putative biomarkers. EHR data analytics typically aims at predicting future outcome based on population and individual longitudinal data. The analytics has a similar process such as data cleaning, clinical features identification, predictive model construction, and clinical validation.
A. Biomedical Big Data
A.1. Big -Omic Data
-Omic data contain a comprehensive catalog of molecular profiles (e.g. genomic, transcriptomic, epigenomic, proteomic, and metabolomic as explained in Table II) in biological samples that provide a basis for precision medicine [17]. The genome, transcriptome, and epigenome are upstream of the proteome and metabolome. A genome is unique and mostly invariant over time with its knowledge embedded in single nucleotide polymorphisms (SNPs), frameshift mutations (insertions or deletions; or indels), copy number variations (CNVs), and other structural variations (SVs) [30, 31]; transcriptomic knowledge is contained in gene expression, transcript expression, gene fusion, and alternative splicing [32, 33]; epigenomic knowledge is carried in protein-DNA binding sites, histone modification patterns, and DNA methylation patterns [34]; proteomic knowledge is reflected by protein expression, post-translational modification, and protein-protein interactions [35]; and metabolomic knowledge is shown in the abundance of metabolites [36]. Because epigenomic information impacts transcriptomic, proteomic, and metabolomic profiles [37], and the proteome and metabolome are directly responsible for the establishment of phenotypes, uncovering interactions among the proteome, metabolome, and upstream processes is a key towards precision medicine.
A.2. Big Electronic Health Record Data
EHR data can be unstructured (e.g., clinical notes) or structured (e.g., ICD-9 diagnosis codes, administrative data, chart, and medication) [38]. Written or dictated clinical notes describe the patient’s condition and are the most efficient and human-intuitive way for clinical documentation. However, they are the most challenging for computer analysis because of (1) unstructured and heterogeneous data formats, (2) abundant typing and spelling errors, (3) violation of natural language grammar, and (4) rich domain-specific abbreviations, acronyms, and idiosyncrasies [39]. Structured EHR data can be categorized into two classes [40]. Administrative data include those remain unchanged during the entire course of a clinical encounter (e.g., demographic data), and those keep updating over time (e.g., diagnoses and procedures) [41]. Ancillary clinical data are frequently recorded during a clinical encounter that can be discrete (e.g., physiological measures, medication, and lab tests), or continuous (e.g., respiration, blood pressure, pulse oximetry, and electrocardiography waveforms captured by sensors, either through bedside monitoring devices or ambulatory, implanted, or wearable devices) [40].
B. Challenges Associated with -Omic and EHR Data
-Omic and EHR big data analytics is challenging due to data frequency; quality; dimensionality; and heterogeneity.
B.1. Diverse Data Collection Frequency
First, different data modalities have different data collection frequency. For example, in -omic data, a genome is invariant over a long period of time, and often only needs a one-time data acquisition, while other types of -omic data vary with environment, tissue types, and time that require multi-time-point acquisition. In EHR, bedside monitoring data are captured at very high frequency, while lab tests may be taken a few times a day. In addition, data generation frequency may be influenced by cost (e.g. proteomic/metabolomic data generated by MS versus genomic/transcriptomic/epigenomic data generated by NGS). Second, data collection frequency can be irregular. For example, in EHR, most clinical variables have irregular sampling frequencies, depending on the criticality of a patient and the easiness of a measurement.
B.2. Inherent Data Quality Issues
In -omic data, quality issues are caused by a combination of biological, instrumental, and environmental factors such as sample contamination [42, 43], batch effects [44, 45], and low signal-to-noise ratios [46, 47]. In EHR data, quality issues include missing data because recorded clinical variables vary with each clinical encounter and depend on clinical team’s assessment of the patient’s condition [48], and erroneous data entries happening due to data entry mistakes or misinterpretation of original documents when entering [49]. For high-resolution waveform data, common quality issues include random noise, gaps in the waveform, and artifacts (e.g., patient’s motion) [50]. These data quality issues may lead to wrong conclusion, but correcting these remains challenging.
B.3. High Dimensionality
A big challenge in either -omic or EHR data mining is the “curse of dimensionality” associated with high-dimensional data [51]. -Omic data often have many dimensions or features (may be more than 104) much larger than the number of samples available, while EHR data may contain a large sample size of high-dimensional data but with each individual sample only sparsely populated. Making sense of these data with statistical significance presents to be challenging.
B.4. Heterogeneous Data Types
In -omics, using underlying molecular fingerprints to characterize disease subtypes may require heterogeneous multi-omic data. For example, the integrative personal omics profile (iPOP) project has integrated multiple molecular expression profiles to uncover dynamic molecular changes between healthy and diseased states [52]. However, integrating multi-omic data is challenging because of variations in represented biological processes, technical and biological noise levels, identification accuracy, spatiotemporal resolution, and many other confounding factors [53]. In EHR, the data are inherently heterogeneous. To accomplish precision medicine, it is necessary and critical to make sense of heterogeneous data.
C. Big Data Analytics for Precision Medicine
C.1. General Analytics for Biomedical Big Data
Most -Omic and EHR are high-dimensional data that not only require longer computational time but also affect the accuracy of analysis. Thus, we try to reduce data dimensionality by identifying a subset of variables or latent factors that preserve as much of the characteristics of the original data as possible with two strategies (Table III): (i) feature selection that aims to select an optimal subset of existing features, and (ii) feature extraction that aims to transform existing features into a more compact set of dimensions [56].
TABLE III.
Selected Methods for Dimensionality Reduction
| Method | Advantages | Limitations |
|---|---|---|
| Feature extraction: PCA, SVD, tensor-based approaches* [54] |
Reduces dimensionality; relatively immune to noise |
Performance usually inferior to supervised approaches; difficult to interpret results |
| Feature selection: filter- based (mRMR), wrapper- based (sequential feature selection)* [55] |
Reduces dimensionality; easy to interpret |
Sometimes affected by noisy data |
Highly impactful method with more than 50,000 relevant papers.
Feature selection techniques consist of filtering, wrapper, or embedded methods. Filtering methods limit the number of features by calculating a score designed to estimate the usefulness of each feature. Thus, they are generally faster and do not require explicit class labeling. The minimum redundancy maximum relevance (mRMR) method is a filtering method that iteratively selects features sharing the most mutual information (relevance) with the least redundancy [57]. In contrast, wrapper methods select a subset of features (i.e. “wrap” the feature selection) for targeted learning models by using evaluation metrics such as cross-validation accuracy [58]. Embedded methods integrate machine learning algorithms (e.g. support vector machines) with recursive feature elimination [59].
Among feature extraction techniques, principal component analysis (PCA) is a basic method that identifies a small number of orthogonal linear vectors [51]. Its performance heavily depends on correctly identifying an optimal number of components, and requires careful testing and validation [60]. Other techniques include artificial neural networks such as autoencoders [61], and nonlinear kernels in PCA [62].
C.2. -Omic Data Pre-processing
NGS and MS high-throughput assays require different pre-processing methods that are summarized in Table IV. NGS is a popular assay for genomic, transcriptomic, and epigenomic studies. Its common pre-processing step is sequence mapping that identifies not only the origin but also the alignment of each read [78]. This step is computationally intensive and requires auxiliary data structures (e.g., the hash table [63] and the Burrows-Wheeler transform [64]), multithreading, or in-memory computing [65] for improved computational efficiency. Genomic studies typically aim to identify variants in a sequenced genome [79]. Small-scale variant (i.e., SNPs and indels) detection uses “per base differences” between reads and the reference genome as the evidence [30, 66]. Large-scale variant (i.e., SVs) detection uses read-pair-based, read-depth-based, split-read-based, and assembly-based methods [80, 81]. Transcriptomic studies mostly center on expression profiling, fusion gene detection, and alternative splicing detection [32, 33]. Expression profiling associates mapped reads with genes and isoforms. Different profiling methods handle multi-mapped reads differently, where some methods associate the reads with all loci [67, 68], while others probabilistically associate the reads with only a few model-inferred loci [69, 70]. Fusion gene detection relies on two factors, the spanning read pairs and the split read [24, 25]. Alternative splicing detection relies on either de novo transcriptome assembly [71, 72, 82–84], or inference from sequence mapping outputs [70, 73]. Epigenomic studies mainly focus on identifying patterns of protein-DNA binding sites, histone modification, and DNA methylation [34]. Epigenomic data pre-processing builds a profile representing the density of reads along the genome, models background noises, and determines statistically significant peaks [78].
TABLE IV.
Selected Tools for -Omic Data Pre-processing
| Tool | Assay | -Omic Data | Key Functionality |
|---|---|---|---|
| GMAP* [63] | Next- generatio n sequenci ng |
Genomic, transcriptomic , and epigenomic |
Sequence mapping |
| BWA* [64] | |||
| STAR* [65] | |||
| GATK* [30] | Genomic | Genomic variant discovery |
|
| SAMtools* [66] | |||
| HTSeq* [67] | Transcriptomi c |
Gene and transcript expression quantification |
|
| BEDTools* [68] | |||
| RSEM* [69] | Gene and transcript expression quantification |
||
| Cufflinks* [70] | |||
| defuse [25] | Gene fusion detection | ||
| TopHat-Fusion [24] | |||
| Trans-ABySS* [71] |
Alternative splicing detection and quantification |
||
| Trinity* [72] | |||
| Cufflinks* [70] | |||
| Scripture* [73] | |||
| MACS* [74] | Epigenomic | ChIP-seq peak calling | |
| SISSRs* [75] | |||
| OpenMS [76] | Mass spectrom etry |
Proteomic and metabolomic |
Peak detection and quantification |
| MZmine 2* [77] |
GMAP stands for genomic mapping and alignment program; BWA, Burrows-Wheeler aligner; STAR, spliced transcripts alignment to a reference; GATK, genome analysis toolkit; RSEM, RNA-seq by expectation-maximization; Trans-ABySS, transcriptome assembly and analysis pipeline; MACS, model-based analysis of ChIP-seq; and SISSRs, site identification from short sequence reads.
Highly impactful tool with more than 50 citations per year.
MS is for proteomic and metabolomic studies, and its pre-processing steps include alignment, baseline correction, and peak detection [85]. In chromatography-coupled MS, chromatographic peak alignment can correct drift to ensure coherent retention time and accurate mass across samples, and mass-to-charge ratio (m/z) alignment can ensure mass spectra and component features are comparable among samples [86]. In MALDI (matrix-assisted laser desorption ionization) MS, baseline correction is particularly important. Low-mass measurement noise from the chemical matrix used in MALDI experiments affects the spectral baseline and needs to be removed prior to analysis [87]. Peak detection is then performed based on criteria such as signal-to-noise ratio, peak shape, and detection thresholds [88]. A common subsequent step is the identification, and potentially the filtering, of isotopic peaks from the spectrum [77].
C.3. Biomarker Identification Using -Omic Data
In practice, different groups of samples are collected for different biological conditions (e.g., disease vs. non-disease) or different time points (e.g., before vs. after a treatment). Thus, Table V summarizes selected tools that identify discriminatory biomarkers among different groups. Most -omic biomarkers are identified by investigating statistically significant differences among groups, such as differentially expressed genes or transcripts [94–96], differential alternative splicing [97, 98], differential protein-DNA binding [99], differential histone modification [100], and differential DNA methylation [101]. The basic idea is to quantify and then fit the abundance of each group to Poisson-based distributions (e.g., the Poisson distribution and the negative binomial distribution), followed by statistical tests (e.g., the Fisher’s exact test and the likelihood ratio test) that determine the statistical significance of each molecular feature. For genomic data, genome-wide association studies (GWAS) uses different approaches (e.g., the chi-squared test or logistic regression) to assess the degree of association between each variant and a targeted trait, and then select most significant variants as biomarkers [105]. Most GWAS focuses on SNP association [89–91], while only a few infer CNV or SV association [92, 93].
TABLE V.
Selected Tools for -Omic Biomarker Identification
| Tool | -Omic Data | -Omic Biomarker | Approach |
|---|---|---|---|
| SNPassoc [89] | Genomic | Significant SNPs associated with traits |
Genome- wide association studies |
| SNPTEST* [90] | |||
| VAT [91] | Significant SNPs and indels associated with traits |
||
| PLINK* [92] | Significant SNPs, indels, and CNVs associated with traits |
||
| CNVRuler [93] | Significant CNVs associated with traits |
||
| edgeR* [94] | Transcriptomic | Differentially expressed genes /transcripts |
Differential analysis (model fitting and statistical tests) |
| DESeq2* [95] | |||
| omniBiomarker [96] | |||
| DiffSplice [97] | Differential alternative splicing |
||
| MATS [98] | |||
| DBChIP [99] | Epigenomic | Differential binding sites | |
| ChIPDiff [100] | Differential histone modification sites |
||
| QDMR [101] | Differentially methylated regions |
||
| DetectTLC [102] | Proteomic and metabolomic |
Molecular patterns in mass spectrometry images |
Similarity scoring |
| Automics [103] | Differentially abundant metabolites |
Supervised and unsupervised learning |
|
| MetaboAnalyst* [104] |
SNPassoc stands for SNP-based whole genome association studies; VAT, variant association tools; PLINK, population-based linkage analyses; edgeR, empirical analysis of digital gene expression data in R; MATS, multivariate analysis of transcript splicing; DBChIP, differential binding with ChIP-seq data; and QDMR, quantitative differentially methylated regions.
Highly impactful tool with more than 50 citations per year.
C.4. Systems Biology Modeling Using -Omic Data
To gain insights about a complex molecular system, we can conduct systems biology modeling using either “static network analysis” or “dynamic temporal analysis” based on -omic features (Table VI).
TABLE VI.
Selected Tools for -Omic Data Modeling
| Tool | Modeling Type |
Approach | Key Functionality |
|---|---|---|---|
| WGCNA* [106] |
Static Network analysis |
Correlation between quantitative variables |
Network construction, module detection, and gene selection |
| CODENSE [107] |
Summary graphs and dense subgraphs for frequent edges |
Mining frequent coherent dense subgraphs across large numbers of massive graphs |
|
| MEMo* [108] | Mutually exclusive genomic alterations |
Network construction, module detection, and gene selection |
|
| CellDesigner [109] |
Dynamic Temporal Analysis |
Ordinary and partial differential equations |
Graphical interface for ODE or PDE model implementation and simulation; systems biology markup language compatibility |
| NetLogo* [110] |
Agent-based models | General-purpose modeling environment capable of simulating hundreds to thousands of interacting agents |
|
| BoolNet [111] | Boolean models | Simulating and analyzing Boolean and probabilistic Boolean models |
|
| Snoopy [112] | Petri nets | Network modeling using Petri nets; hierarchical structure and multiple class compatibility |
WGCNA stands for weighted correlation network analysis; CODENSE, coherent dense subgraphs; and MEMo, mutual exclusivity modules in cancer.
Highly impactful tool with more than 50 citations per year.
Static network analysis studies the interactome (i.e., a complete set of molecular interactions) with three steps [113]: identifying a network scaffold that describes interactions among -omic features [106, 108]; decomposing the network scaffold into smaller network modules [106–108]; and mathematically representing each network module for downstream simulation and analysis [114]. Most interactome networks use a single -omic data such as metabolic networks and gene regulatory networks. Few incorporate multi-omic data but are limited to simpler organisms (e.g., S. cerevisiae and C. elegans) [115].
Dynamic temporal analysis (e.g., ordinary or partial differential equations, Boolean networks, agent-based models, and Petri nets [116]) makes use of temporal measurement of - omic data to develop and validate dynamic predictive models of complex systems. For example, a recent study on A. thaliana used a Granger causality model to integrate two types of metabolomic data acquired at multiple time points for studying the interaction of primary and secondary metabolism [117].
C.5. EHR Data Pre-processing
Information embedded in EHR is abundant but disorganized in nature. Thus, EHR data requires systematic pre-processing that are summarized in Table VII. On EHR missing data, conventional approaches either impute missing values by the mean or median in a population, or list-wise or pair-wise delete records with missing values. These approaches are simple and easy to implement, but they ignore the underlying data structure and tend to introduce additional biases [128]. Thus, more robust missing data imputation methods such as interpolation [129], multiple imputation [130], expectation maximization [131], and maximum likelihood [132] are needed.
TABLE VII.
Selected Methods for EHR Data Pre-processing
| Method | Advantages | Limitations |
|---|---|---|
| Missing data: list-wise deletion, mean filling* [118, 119] |
Simple to implement; complete case analysis |
Loss of statistical power; introduces biases; underestimates variances |
| Missing data: hot deck, nearest neighbor* [120] |
Simple to implement and interpret; immune to cross-user inconsistencies |
Introduces biases; underestimates variances |
| Missing data: interpolation (linear, piece-wise linear, spline, cubic) [121] |
Simple to implement and interpret; direct estimation on the basis of neighbors |
Does not account for relationships among different features |
| Missing data: model-based filling (expectation maximization, maximum likelihood, multiple imputations)* [122] |
Accounts for uncertainty in imputations |
Does not account for missing data mechanisms (i.e., MCAR, MAR, and MNAR) |
| Waveforms: noise filtering (IIR, FIR, PCA, ICA, Kalman filter, wavelets) [50, 123] |
Generally simple to implement |
Falls short in situations where “true” waveform is obscured by artifact such as patient motion |
| Waveforms: signal quality indices [123–125] |
Human-interpretable metrics of signal quality |
Can be complex to implement and computationally intensive; may require ad-hoc calibration based on the features of the target waveform |
| Waveforms: sensor fusion [126, 127] |
Improved SNR; reduces data dimensionality while increasing data quality |
Computationally intensive; loss of detail from individual sensor waveforms |
Highly impactful method with more than 50,000 relevant papers.
On high time-resolution waveform data quality issues [50], we can use (i) filtering strategies such as median filtering, Kalman filtering, and model-based filtering to handle noise [50, 123]; (ii) signal quality indices that detect the presence of expected physiological features, quantify the agreement between signals with mutual information, or infer other ad hoc definitions of signal quality to identify artifacts and gaps in the waveform [123–125]; and (iii) sensor fusion techniques (e.g. using redundant measurements of electrocardiography, blood pressure sensors, and photoplethysmography to derive a more reliable measure of heart rate than any single signal alone [126, 127]) to correct artifacts and fill in gaps in the waveform.
C.6. EHR Data Mining
To derive actionable knowledge from complex EHR big data, two strategies such as static endpoint prediction and temporal data mining are summarized in Table VIII.
TABLE VIII.
Selected Methods for EHR Data Mining
| Method | Advantages | Limitations |
|---|---|---|
| Logistic regression, cox regression, local regression (LOESS)* [133] |
Simple to implement and interpret; direct estimates of relevant hazards for Cox regression |
Sensitive to outliers |
| Logistic regression with LASSO regularization [134] |
Reduces feature space | Prone to overfitting |
| Hidden Markov models [135] |
Simultaneous detection, segmentation, and classification in a waveform |
Sensitive to the design of the Markov model being trained |
| Conditional random fields [136] |
Supports temporal analysis; resistant to differences in class prevalence |
Sensitive to regularization and feature space size |
| Relational subgroup discovery, episode rule mining, windowing* [137] |
Valid sequential techniques for some clinical applications |
Tradeoffs between simplicity, complexity, and temporal resolution |
| Rule mining, Allen’s interval algebra, directed acyclic graph* [138] |
Temporal mining/modeling capabilities |
Requires specific experimental design |
Highly impactful method with more than 50,000 relevant papers.
C.6.1. Static Endpoint Prediction
After dimensionality reduction, we can model the relationship between selected clinical features (i.e. the patient’s condition) and targeted clinical endpoints (i.e. the clinical outcome) with three groups of techniques: Regression analysis is a statistical process that estimates the relationship between independent variables (i.e., features) and dependent variables (i.e., endpoints). If dependent variables follow distributions such as normal, Poisson, and binomial, we can use a generalized linear model for regression model fitting; Classification involves building statistical models that assign a new observation to a known class. Many classification techniques such as decision trees, k-nearest neighbors, and support vector machines (SVM) prove to be effective in clinical applications; Associate Rule Learning (ARL) discovers frequent and reliable associations among clinical variables, and these association rules describe that if all elements in the antecedent occurs, all elements in the consequent should occur with certain confidence [28]. In general, these machine learning techniques prefer a large sample size.
C.6.2. Temporal Data Mining
EHR captures diagnosis, treatment, and outcome chronologically throughout a medical encounter, and thus, it is important to model temporal relationship between events using temporal data mining techniques such as the hidden Markov model (HMM) and the conditional random field (CRF) [135, 136]. One constraint of HMM and CRF is that they require predefined clinical variables and outcome categories ofen difficult to generalize for a given treatment of a given patient. Thus, temporal association rule mining (TARM) is proposed to discover causality between the event and outcome. A temporal association rule, denoted by A →T C, describes an antecedent A followed by a consequent C separated by a time difference T. Because the selection of the event and outcome is flexible, TARM model can be tailored for any event-outcome combination in various clinical settings [137, 139].
D. Enablers of Biomedical Big Data Analytics
The big data revolution has led to the development of enterprise tools and platforms for extracting, summarizing, and interpreting knowledge from rapidly generated data, for business intelligence, analytics, and predictive modeling as summarized in Table IX [11, 140, 141].
TABLE IX.
Selected Platforms for Big Data Analytics
| Platform | Advantages | Limitations |
|---|---|---|
| Apache Hadoop (MapReduce)* [11, 142] |
Horizontally scalable; fault- tolerant; designed to be deployed on commodity-grade hardware; free and open-source |
Generally most effective for batch-mode processing; not always appropriate for real- time, online analytics |
| IBM InfoSphere Platform* [143] |
Includes purpose-built tools to handle streaming information; integrates with open-source tools such as Hadoop |
Commercial licensing |
| Apache Spark Streaming* [144] |
Integrates with the Hadoop stack; allows one code base for both batch-mode and online analysis |
Depends on more expensive hardware with large amounts of RAM to work efficiently |
| Tableau, QlikView, TIBCO Spotfire, and other visual analytics tools* |
Visualization of large and complex data sets |
Generally incomplete solutions, requiring other tools to effectively handle data storage |
Highly impactful platform.
Distributed computing systems such as Apache Hadoop (based on MapReduce) provide the storage and processing backbone for dealing with very large datasets [142]. Specific tools also exist to solve more specialized problems. For example, IBM InfoSphere Streams and Apache Spark Streaming can handle real-time streaming data [143, 144]. Cloud computing providers such as Amazon Elastic Compute Cloud (EC2) can provide on-demand computing power to accommodate scalable growth from development to truly big data production [145]. Many cloud-based services in bioinformatics such as Illumina’s BaseSpace [146] and the Galaxy project [147] are deployed on Amazon EC2.
To deploy biomedical big data for precision medicine in health care, there is a critical need to address domain-specific challenges such as the requirements of HIPAA (Health Insurance Portability and Accountability Act), HITECH (Health Information Technology for Economic and Clinical Health), and other privacy regulations. Thus, security is an important enabling technology (e.g., encryption for protected health information) in biomedical big data [140, 145, 148].
III. Case Studies
In this section, we present two real-world applications to illustrate the utility of biomedical big data analytics for precision medicine: (1) integrative -omic data for the improved understanding of cancer mechanisms (see Fig. 2), and (2) the incorporation of genomic knowledge into the EHR system for improved patient diagnosis and care (see Fig. 3).
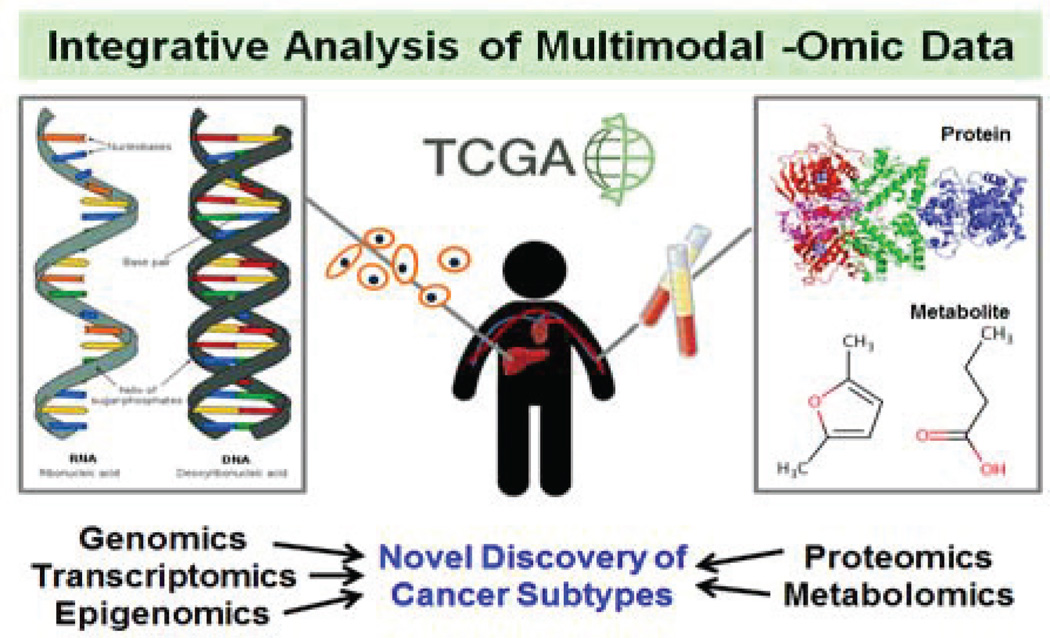
Fig. 2.
Integrative analysis of multi-omic data leads to the improved understanding of cancer mechanisms, which in turn enables more precise classification of cancer subtypes.
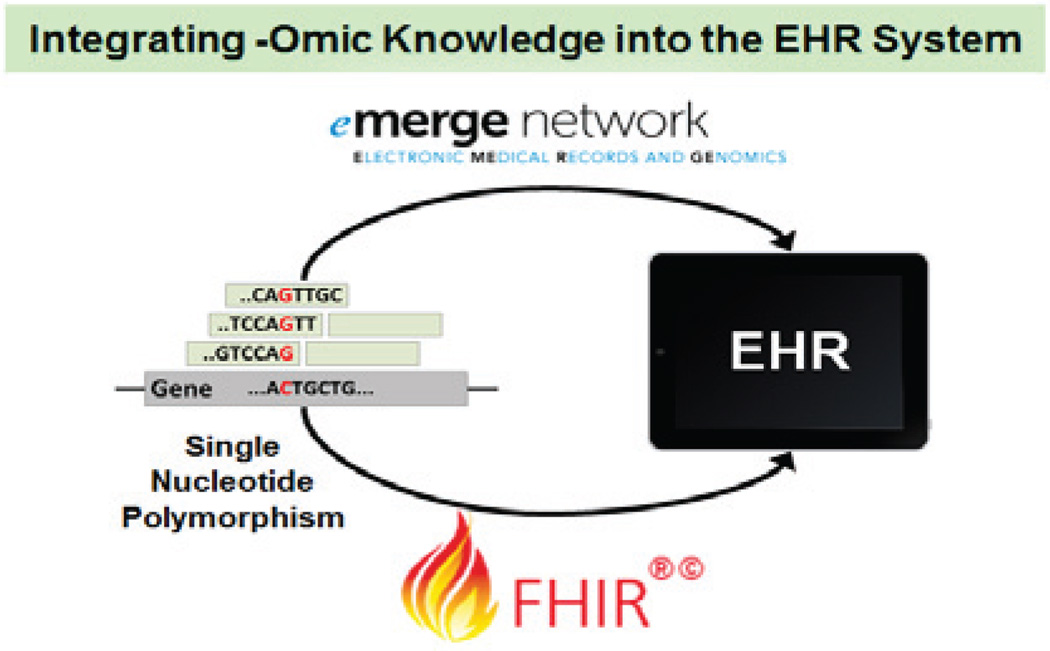
Fig. 3.
Integrating derived -omic knowledge into the existing EHR system is an approach to utilizing molecular information for clinical decision support, and it also help deliver precision medicine.
A. Integrative -Omics for Precise Cancer Understanding
One notable effort that integrates multi-omic data for the improved understanding of cancer mechanisms is The Cancer Genome Atlas (TCGA) [149]. TCGA hosts public datasets of 27 cancer types with more than 11,000 patient cases. Each patient is annotated with clinical data (i.e. demographic, diagnostic, and survival data) and multimodal -omic data (i.e., genomic, transcriptomic, epigenomic, and proteomic).
We use head and neck squamous cell carcinoma (HNSCC) as an example to illustrate the integrative multi-omic study for precision medicine [150]. In 2014, a pan-cancer study with twelve cancer types using multi-omic TCGA data was performed [151]. Among 3,527 samples in total, 305 were HNSCC. Six different data types (i.e. DNA copy number, methylation, mutation, and expression of mRNAs, miRNAs, and proteins) were analyzed both separately and integratively. By using clustering-based methods, pathway activities (inferred from gene expression and copy number data) have shown common copy number variations, mutation frequency patterns, and survival patterns between HNSCC and lung squamous cell carcinomas or some bladder cancers. Such integrative pan-cancer analysis provides more precise subtyping across multiple cancers sharing common molecular-level processes underlying cancer development. This new subtyping system reflects precision medicine because it finds precise classification of patients into disease subgroups.
TCGA Research Network has published more than 30 articles describing multi-omic investigation on numerous cancer types, and identified more precise, clinically relevant subtyping for multiple cancers [152–154].
B. Adoption of Genomics in EHR for Precision Medicine
In a clinical setting, healthcare providers use electronic medical record (EMR) for clinical decision support. Thus, it is important to incorporate -omic data and knowledge into EMR. The Electronic Medical Records and Genomics (eMERGE) Network consortium aims to identify causal genomic variants (mostly SNPs) for EMR-based phenotypes and to integrate identified genotype-phenotype associations into the EMR system [155]. One crucial challenge is on how to store variants present in an individual or even in family members in the EMR [156]. The consortium has proposed several recommendations on augmenting the current EMR structure: (1) it should store various genomic variants, such as SNPs, indels, and CNVs, in a discrete computable format; (2) it needs to satisfy interoperability to reduce the burden in data transfer and update within and between healthcare facilities; (3) it has to support rule-based decision support engines; and (4) it must contain abundant visualization elements for easier interpretation [157]. Another big challenge is that each individual typically has millions of variants. The consortium has proposed one potential solution that stores only known pathological variants in the EMR system. However, because the set of known pathological variants may change over time, this approach may lead to the inclusion of false positive and the exclusion of false negative variants. Thus, an alternative solution is to archive raw data in separate repositories easily accessible when necessary [158].
EMR is only for local clinic and hospital, while EHR contains and shares medical records among all participant clinics and hospitals [159]. Thus, interoperability is critical in using big data for precision medicine. Recently, the Health Level Seven International (HL7) proposed the Fast Healthcare Interoperability Resources (FHIR) standard that addresses this important issue. On clinical genomics, several new FHIR resources and extension definitions are designed for variant data [160]. With such the standardized data exchange protocol, clinicians can utilize genomic information with other existing EHR data to determine the most effective treatment for each patient, which is a paradigm shift towards precision medicine.
IV. Biomedical Big Data Initiatives
To apply big data analytics for precision medicine, Table X summarizes multiple consortium initiatives that collect and organize data from various projects and trials, and make them available to the research community for secondary data use.
TABLE X.
Selected Biomedical Big Data Initiatives
| Consortium | Data Sources | Data Elements |
|---|---|---|
| TCGA | Multi-omic data for 27 cancer types, covering more than 11,000 cases |
Clinical, genomic, transcriptomic, epigenomic, and proteomic data |
| Project Data Sphere |
Patient-level data from comparator arms of Phase IIB and III clinical trials; currently contains 33 trials covering 12 cancer types |
Common data include baseline, safety, efficacy, medication, dosing, lab test, medical history, and demographic data |
| TARGET | Multi-omic data for 7 types of childhood cancers |
Clinical, genomic, transcriptomic, and epigenomic data |
| 1,000 Genomes Project |
Large-scale genome sequencing project for populations of African, European, and East Asian ancestry |
Low-coverage whole genome sequencing for 179 individuals; high-coverage targeted exome sequencing for 697 individuals |
| 100,000 Genomes Project |
Large-scale genome sequencing project for studying cancers and rare diseases in the UK |
Genome sequencing will be completed in 2017 |
| ICGC | Genomic data for 18 cancer types; partially overlap the TCGA data |
SNPs, CNVs, methylation, and gene and miRNA expression |
| RD- Connect |
Infrastructure project funded by European Union for facilitating rare disease research |
Currently links to 3 biobanks and more than 150 rare disease registries |
| ADNI | Multi-center, longitudinal study with elderly control subjects, early Alzheimer’s disease subjects, and mild cognitive impairment subjects |
Clinical, genetic, magnetic resonance imaging, and positron emission tomography imaging data |
| iDASH | Data from 17 focused trials, each of which represents a specific objective and a patient population |
Imaging, EHR, sensor, and genomic data from multiple clinical trials |
| GHO | Worldwide population and environmental data for infectious diseases, noncommunicable diseases, sexually transmitted diseases, and children’s health |
Population-level statistics and modeling |
| BMIC | Large trials encompassing thousands of samples |
EHR, imaging, genetic, and social research data |
| MIMIC II | ICU data for more than 30,000 patients with more than 40,000 ICU stays |
Chart data, administrative data, alert data, lab results, electronic documentation, and bedside monitor trends and waveforms |
| HIW | Federal data for aggregated health indices by geography; covers data from claims, healthcare cost, to population statistics |
Data element varies, depending on the trials |
TCGA stands for The Cancer Genome Atlas; TARGET, Therapeutically Applicable Research to Generate Effective Treatments; ICGC, International Cancer Genome Consortium; ADNI, Alzheimer’s Diseases Neuroimaging Initiative; iDASH, Integrating Data for Analysis, Anonymization, and Sharing; GHO, WHO Global Health Observatory Data Repository; BMIC, Trans-NIH BioMedical Informatics Coordinating Committee; MIMIC II, Multiparameter Intelligent Monitoring in Intensive Care II; and HIW, Health Indicators Warehouse.
First, initiatives such as Project Data Sphere aim to improve research efficiency and to encourage collaboration by integrating information of clinical trials for different cancers. For example, the European Union-funded RD-Connect aggregates data of multiple rare diseases from around the world. The Cancer Genome Atlas (TCGA) of US, Therapeutically Applicable Research to Generate Effective Treatments (TARGET), and the International Cancer Genome Consortium (ICGC) aim to study multiple aspects of individual diseases by collecting multi-omic data of hundreds of patients for each cancer type. In contrast, the US 1,000 Genomes Project and the UK-based 100,000 Genomes Project aim to connect genotypes with phenotypes using single -omic data type.
Second, large data repositories have been created and maintained by organizations such as US National Institutes of Health (NIH) and the World Health Organization (WHO). The Trans-NIH BioMedical Informatics Coordinating Committee (BMIC) has established a data repository that archives the data from 61 large multi-center studies for promoting secondary use of biomedical data [161]. The Global Health Observatory Data Repository (GHO) is maintained by WHO for population-level health studies [162]. As another example, the Health Indicators Warehouse (HIW) within the US Department of Health and Human Services provides country-level and state-level aggregated clinical information [163].
V. Discussion
Among many data types included in the NIH Big Data to Knowledge (BD2K) Initiative, -omic data, EHR, and medical imaging data are the three most important biomedical big data. We conducted the review of -omic and EHR data because of their close relationship with precision medicine [3, 5, 17, 18].
Big data have had major societal impact in energy, environment, financial, and others. They motivate rapid advances in data storage, data mining and analytics, data retrieval, and data visualization [164, 165]. When applying to biomedicine and healthcare, big data will improve quality and outcome by (i) discovering new knowledge (e.g., automated identification of postoperative complications in EHR data [166]); (ii) disseminating new knowledge (e.g., data-driven clinical decision support systems such as IBM Watson); (iii) incorporating -omic data into EHR (e.g. eMERGE network [167]); and (iv) implementing patient-centered care (e.g., e-health [168]).
To accelerate the delivery of precision medicine, more research is needed in the following biomedical big data areas:
-Omic Data Integration: As illustrated by the TCGA case study, integrative multi-omic data analysis is of growing importance because it provides holistic view of molecular fingerprints for each patient’s condition. Recent research has shown positive impact of knowledge and insight obtained from integrative analysis of genomic and transcriptomic [169], transcriptomic and proteomic [170], and multiple -omic data types [53, 151] on disease diagnosis, prognosis, and treatment. The next important direction is the development of guidelines (or best practices) for -omic data integration and interpretation that will in turn enable better prediction of bio-system behavior, and safer and more effective therapeutics.
Waveform and Irregularly Spaced Time Series Analysis: Real-time streaming data analytics needs to be further developed due to the pervasive use of wearable sensors in either the critical care setting or in the continuous home monitoring setting for fitness and preventative medicine [171], and the need to reduce alarm fatigue [172]. However, the challenge for irregularly sampled temporal data remains and requires advanced imputation techniques and robust parameter extraction techniques [50, 173].
Patient Similarity: Precision medicine promotes precise subgroup classification of patients based on biological basis such as molecular profiles. Thus, EHR mining can assist in patient classification based on clinical measurements (e.g. drug responses, physiological signals, and disease susceptibility). However, because of high patient variability for any disease, the precise subtypes of many diseases remain unknown as of today, and it requires systematic big data analytics to model physician knowledge to validate the reliability of patient subgrouping based on EHR mining.
VI. Conclusion
In this review, we present -omic and EHR big data challenges and current progressed. We provide case studies to show how big data analytics can facilitate precision medicine. Because biomedical big data analytics is in its infancy, more biomedical data scientists and engineers are needed to gain necessary biomedical knowledge, to use large data provided by biomedical big data initiatives, and to put concerted effort in areas such as multi-omic data integration, waveform and time series data analysis, and patient similarity and so on to speed up big data research for precision medicine. By delivering the most suitable and effective treatment to each patient based on their precise subtyping information, the healthcare system can achieve better care efficiency and quality.
Acknowledgments
This work was supported in part by grants from the National Center for Advancing Translational Sciences of the National Institutes of Health (NIH) under Award Number UL1TR000454, NIH R01CA163256, the Georgia Research Alliance Cancer Coalition (Distinguished Cancer Scholar Award to Professor May D. Wang), the Children’s Healthcare of Atlanta, Centers for Disease Control and Prevention, Microsoft Research, and the Hewlett-Packard.
Footnotes
Disclaimer
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Po-Yen Wu, School of Electrical and Computer Engineering, Georgia Institute of Technology, Atlanta, GA 30332, USA.
Chih-Wen Cheng, Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA, USA.
Chanchala D. Kaddi, Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA, USA
Janani Venugopalan, Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA, USA.
Ryan Hoffman, Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA, USA.
May D. Wang, Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA, USA.
References
- 1.Fernald GH, et al. Bioinformatics challenges for personalized medicine. Bioinformatics. 2011 Jul;27:1741–1748. doi: 10.1093/bioinformatics/btr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hood L, Friend SH. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat Rev Clin Oncol. 2011 Mar;8:184–187. doi: 10.1038/nrclinonc.2010.227. [DOI] [PubMed] [Google Scholar]
- 3.Desmond-Hellmann S, et al. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington (DC): The National Academies Press; 2011. [PubMed] [Google Scholar]
- 4.Katsnelson A. Momentum grows to make 'personalized' medicine more 'precise'. Nature Medicine. 2013 Mar;19:249. doi: 10.1038/nm0313-249. [DOI] [PubMed] [Google Scholar]
- 5.Mirnezami R, et al. Preparing for Precision Medicine. New Engl J Med. 2012 Feb;366:489–491. doi: 10.1056/NEJMp1114866. [DOI] [PubMed] [Google Scholar]
- 6.Chute CG, et al. Some experiences and opportunities for big data in translational research. Genet Med. 2013 Oct;15:802–809. doi: 10.1038/gim.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai L, et al. Bioinformatics clouds for big data manipulation. Biology Direct. 2012 Nov;7 doi: 10.1186/1745-6150-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marx V. Biology: The big challenges of big data. Nature. 2013 Jun;498:255–260. doi: 10.1038/498255a. [DOI] [PubMed] [Google Scholar]
- 9.O'Driscoll A, et al. 'Big data', Hadoop and cloud computing in genomics. J Biomed Inform. 2013 Oct;46:774–781. doi: 10.1016/j.jbi.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Murdoch TB, Detsky AS. The Inevitable Application of Big Data to Health Care. J Amer Med Assoc. 2013 Apr;309:1351–1352. doi: 10.1001/jama.2013.393. [DOI] [PubMed] [Google Scholar]
- 11.Raghupathi W, Raghupathi V. Big data analytics in healthcare: promise and potential. Health Inf Sci Syst. 2014 Feb;2 doi: 10.1186/2047-2501-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bates DW, et al. Big Data In Health Care: Using Analytics To Identify And Manage High-Risk And High-Cost Patients. Health Affairs. 2014 Jul;33:1123–1131. doi: 10.1377/hlthaff.2014.0041. [DOI] [PubMed] [Google Scholar]
- 13.Hsu W, et al. Biomedical imaging informatics in the era of precision medicine: progress, challenges, and opportunities. J Am Med Inform Assn. 2013 Nov;20:1010–1013. doi: 10.1136/amiajnl-2013-002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013 Dec;26:1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banaee H, et al. Data Mining for Wearable Sensors in Health Monitoring Systems: A Review of Recent Trends and Challenges. Sensors. 2013 Dec;13:17472–17500. doi: 10.3390/s131217472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu LD, et al. Internet of Things in Industries: A Survey. IEEE T Ind Inform. 2014 Nov;10:2233–2243. [Google Scholar]
- 17.Collins FS, Varmus H. A New Initiative on Precision Medicine. New Engl J Med. 2015 Feb;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohane IS. Ten things we have to do to achieve precision medicine. Science. 2015 Jul;349:37–38. doi: 10.1126/science.aab1328. [DOI] [PubMed] [Google Scholar]
- 19.Hillestad R, et al. Can electronic medical record systems transform health care? Potential health benefits, savings, and costs. Health Affairs. 2005 Sep;24:1103–1117. doi: 10.1377/hlthaff.24.5.1103. [DOI] [PubMed] [Google Scholar]
- 20.NHGRI. Available: https://www.genome.gov/10000202/fact-sheets/
- 21.Romero R, et al. The use of high-dimensional biology (genomics, transcriptomics, proteomics, and metabolomics) to understand the preterm parturition syndrome. Bjog-an International Journal of Obstetrics and Gynaecology. 2006 Dec;113:118–135. doi: 10.1111/j.1471-0528.2006.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nature. Available: http://www.nature.com/scitable. [Google Scholar]
- 23.Feuk L, et al. Structural variation in the human genome. Nature Reviews Genetics. 2006 Feb;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 24.Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biology. 2011 Aug;12 doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McPherson A, et al. deFuse: An Algorithm for Gene Fusion Discovery in Tumor RNA-Seq Data. Plos Comput Biol. 2011 May;7 doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annual Review of Biochemistry. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 27.What-is-Epigenetics. Available: http://www.whatisepigenetics.com/ [Google Scholar]
- 28.Cheng C-W, et al. icuARM-An ICU Clinical Decision Support System Using Association Rule Mining. IEEE J Transl Eng Heal Med. 2013 Nov;1 doi: 10.1109/JTEHM.2013.2290113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.H. L. S. International. Available: http://hl7.org/fhir/ [Google Scholar]
- 30.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011 May;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie C, Tammi MT. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics. 2009 Mar;10 doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nature Reviews Genetics. 2011 Feb;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols. 2012 Mar;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirst M, Marra MA. Next generation sequencing based approaches to epigenomics. Brief Funct Genomics. 2010 Dec;9:455–465. doi: 10.1093/bfgp/elq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003 Mar 13;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 36.Buchholz A, et al. Metabolomics: quantification of intracellular metabolite dynamics. Biomolecular Engineering. 2002 Jun;19:5–15. doi: 10.1016/s1389-0344(02)00003-5. [DOI] [PubMed] [Google Scholar]
- 37.Liu SJ. Epigenetics advancing personalized nanomedicine in cancer therapy. Adv Drug Deliver Rev. 2012 Oct;64:1532–1543. doi: 10.1016/j.addr.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayrinen K, et al. Definition, structure, content, use and impacts of electronic health records: A review of the research literature. International Journal of Medical Informatics. 2008 May;77:291–304. doi: 10.1016/j.ijmedinf.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 39.Meystre SM, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008;35:128–144. [PubMed] [Google Scholar]
- 40.Jensen PB, et al. Mining electronic health records: towards better research applications and clinical care. Nature Reviews Genetics. 2012 Jun;13:395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 41.Romano PS, et al. A comparison of administrative versus clinical data: coronary artery bypass surgery as an example. J Clin Epidemiol. 1994 Mar;47:249–260. doi: 10.1016/0895-4356(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 42.Patel RK, Jain M. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. Plos One. 2012 Feb 1;7 doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stokes TH, et al. chip artifact CORRECTion (caCORRECT): a bioinformatics system for quality assurance of genomics and proteomics array data. Ann Biomed Eng. 2007 Jun;35:1068–1080. doi: 10.1007/s10439-007-9313-y. [DOI] [PubMed] [Google Scholar]
- 44.Leek JT, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010 Oct;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson WE, et al. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007 Jan;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 46.Ledergerber C, Dessimoz C. Base-calling for next-generation sequencing platforms. Briefings in Bioinformatics. 2011 Sep;12:489–497. doi: 10.1093/bib/bbq077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith CA, et al. XCMS: Processing mass spectrometry data for metabolite profiling using Nonlinear peak alignment, matching, and identification. Analytical Chemistry. 2006 Feb 1;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 48.Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc. 2013 Jan;20:144–151. doi: 10.1136/amiajnl-2011-000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bowman S. Impact of electronic health record systems on information integrity: quality and safety implications. Perspect Health Inf Manag. 2013 Oct;10 [PMC free article] [PubMed] [Google Scholar]
- 50.Clifford GD, et al. Robust parameter extraction for decision support using multimodal intensive care data. Philos T Roy Soc A. 2009 Jan;367:411–429. doi: 10.1098/rsta.2008.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Girolami M, et al. Analysis of complex, multidimensional datasets. Drug Discovery Today: Technologies. 2006 Apr;3:13–19. doi: 10.1016/j.ddtec.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 52.Chen R, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012 Mar;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stanberry L, et al. Integrative Analysis of Longitudinal Metabolomics Data from a Personal Multi-Omics Profile. Metabolites. 2013 Sep;3:741–760. doi: 10.3390/metabo3030741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luukka P, Lampinen J. A Classification Method Based on Principal Component Analysis and Differential Evolution Algorithm Applied for Prediction Diagnosis from Clinical EMR Heart Data Sets. Computational Intelligence in Optimization: Applications and Implementations. 2010;7:263–283. [Google Scholar]
- 55.Huang Y, et al. Feature selection and classification model construction on type 2 diabetic patients' data. Artif Intell Med. 2007 Nov;41:251–262. doi: 10.1016/j.artmed.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 56.Cunningham P. Dimension Reduction. Machine Learning Techniques for Multimedia: Case Studies on Organization and Retrieval. 2008:91–112. [Google Scholar]
- 57.Peng H, et al. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE T Pattern Anal. 2005 Aug;27:1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 58.Kohavi R, John GH. Wrappers for feature subset selection. Artificial Intelligence. 1997 Dec;97:273–324. [Google Scholar]
- 59.Saeys Y, et al. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007 Oct;23:2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- 60.Coste Jl, et al. Methodological issues in determining the dimensionality of composite health measures using principal component analysis: Case illustration and suggestions for practice. Quality of Life Research. 2005 May;14:641–654. doi: 10.1007/s11136-004-1260-6. [DOI] [PubMed] [Google Scholar]
- 61.Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006 Jul;313:504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 62.Scholkopf B, et al. Nonlinear component analysis as a kernel eigenvalue problem. Neural Computation. 1998 Jul;10:1299–1319. [Google Scholar]
- 63.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005 May;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 64.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009 Jul;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013 Jan;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011 Nov;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anders S, et al. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015 Jan;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010 Mar;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011 Aug;12 doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010 May;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robertson G, et al. De novo assembly and analysis of RNA-seq data. Nature Methods. 2010 Nov;7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 72.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology. 2011 Jul;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nature Biotechnology. 2010 May;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biology. 2008 Sep;9 doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jothi R, et al. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Research. 2008 Sep;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sturm M, et al. OpenMS - an open-source software framework for mass spectrometry. BMC Bioinformatics. 2008 Mar;9 doi: 10.1186/1471-2105-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pluskal T, et al. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010 Jul;11 doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pepke S, et al. Computation for ChIP-seq and RNA-seq studies. Nature Methods. 2009 Nov;6:S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pabinger S, et al. A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform. 2014 Mar;15:256–278. doi: 10.1093/bib/bbs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alkan C, et al. Genome structural variation discovery and genotyping. Nature Reviews Genetics. 2011 May;12:363–375. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao M, et al. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinformatics. 2013 Sep;14 doi: 10.1186/1471-2105-14-S11-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chang Z, et al. Bridger: a new framework for de novo transcriptome assembly using RNA-seq data. Genome Biology. 2015 Feb;16 doi: 10.1186/s13059-015-0596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology. 2015 Mar;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martin JA, Wang Z. Next-generation transcriptome assembly. Nature Reviews Genetics. 2011 Oct;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 85.Mueller LN, et al. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J Proteome Res. 2008 Jan;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 86.Dunn WB, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols. 2011 Jun;6:1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 87.Alexandrov T. MALDI imaging mass spectrometry: statistical data analysis and current computational challenges. BMC Bioinformatics. 2012 Nov;13 doi: 10.1186/1471-2105-13-S16-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang C, et al. Comparison of public peak detection algorithms for MALDI mass spectrometry data analysis. BMC Bioinformatics. 2009 Jan;10 doi: 10.1186/1471-2105-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gonzalez JR, et al. SNPassoc: an R package to perform whole genome association studies. Bioinformatics. 2007 Mar;23:644–645. doi: 10.1093/bioinformatics/btm025. [DOI] [PubMed] [Google Scholar]
- 90.Marchini J, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genetics. 2007 Jul;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 91.Wang GT, et al. Variant Association Tools for Quality Control and Analysis of Large-Scale Sequence and Genotyping Array Data. Am J Hum Genet. 2014 May;94:770–783. doi: 10.1016/j.ajhg.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim JH, et al. CNVRuler: a copy number variation-based case-control association analysis tool. Bioinformatics. 2012 Jul;28:1790–1792. doi: 10.1093/bioinformatics/bts239. [DOI] [PubMed] [Google Scholar]
- 94.Robinson MD, et al. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010 Jan;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Love MI, et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014 Dec;15 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Phan JH, et al. omniBiomarker: A Web-Based Application for Knowledge-Driven Biomarker Identification. Ieee Transactions on Biomedical Engineering. 2013 Dec;60:3364–3367. doi: 10.1109/TBME.2012.2212438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hu Y, et al. DiffSplice: the genome-wide detection of differential splicing events with RNA-seq. Nucleic Acids Research. 2013 Jan;41 doi: 10.1093/nar/gks1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shen SH, et al. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Research. 2012 Apr;40 doi: 10.1093/nar/gkr1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liang K, Keles S. Detecting differential binding of transcription factors with ChIP-seq. Bioinformatics. 2012 Jan;28:121–122. doi: 10.1093/bioinformatics/btr605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu H, et al. An HMM approach to genome-wide identification of differential histone modification sites from ChIP-seq data. Bioinformatics. 2008 Oct;24:2344–2349. doi: 10.1093/bioinformatics/btn402. [DOI] [PubMed] [Google Scholar]
- 101.Zhang Y, et al. QDMR: a quantitative method for identification of differentially methylated regions by entropy. Nucleic Acids Research. 2011 May;39 doi: 10.1093/nar/gkr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kaddi CD, et al. DetectTLC: Automated Reaction Mixture Screening Utilizing Quantitative Mass Spectrometry Image Features. J Am Soc Mass Spectrom. 2015 Oct; doi: 10.1007/s13361-015-1293-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang T, et al. Automics: an integrated platform for NMR-based metabonomics spectral processing and data analysis. BMC Bioinformatics. 2009 Mar;10 doi: 10.1186/1471-2105-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xia J, et al. MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic Acids Research. 2012 Jul;40:W127–W133. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bush WS, Moore JH. Chapter 11: Genome-Wide Association Studies. Plos Comput Biol. 2012 Dec;8 doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008 Dec;9 doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hu HY, et al. Mining coherent dense subgraphs across massive biological networks for functional discovery. Bioinformatics. 2005 Jun;21:I213–I221. doi: 10.1093/bioinformatics/bti1049. [DOI] [PubMed] [Google Scholar]
- 108.Ciriello G, et al. Mutual exclusivity analysis identifies oncogenic network modules. Genome Research. 2012 Feb;22:398–406. doi: 10.1101/gr.125567.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Funahashi A, et al. CellDesigner 3.5: A versatile modeling tool for biochemical networks. Proceedings of the IEEE. 2008 Aug;96:1254–1265. [Google Scholar]
- 110.Wilensky U. NetLogo. 1999 Available: http://ccl.northwestern.edu/netlogo/ [Google Scholar]
- 111.Mussel C, et al. BoolNet—an R package for generation, reconstruction and analysis of Boolean networks. Bioinformatics. 2010 May;26:1378–1380. doi: 10.1093/bioinformatics/btq124. [DOI] [PubMed] [Google Scholar]
- 112.Rohr C, et al. Snoopy—a unifying Petri net framework to investigate biomolecular networks. Bioinformatics. 2010 Apr;26:974–975. doi: 10.1093/bioinformatics/btq050. [DOI] [PubMed] [Google Scholar]
- 113.Joyce AR, Palsson BO. The model organism as a system: integrating 'omics' data sets. Nat Rev Mol Cell Bio. 2006 Mar;7:198–210. doi: 10.1038/nrm1857. [DOI] [PubMed] [Google Scholar]
- 114.Barabasi AL, et al. Network medicine: a network-based approach to human disease. Nature Reviews Genetics. 2011 Jan;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vidal M, et al. Interactome Networks and Human Disease. Cell. 2011 Mar;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.de Jong H. Modeling and Simulation of Genetic Regulatory Systems: A Literature Review. J Comput Biol. 2002 Jul;9:67–103. doi: 10.1089/10665270252833208. [DOI] [PubMed] [Google Scholar]
- 117.Doerfler H, et al. Granger causality in integrated GC-MS and LCMS metabolomics data reveals the interface of primary and secondary metabolism. Metabolomics. 2013 Jun;9:564–574. doi: 10.1007/s11306-012-0470-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee J, et al. Interrogating a clinical database to study treatment of hypotension in the critically ill. BMJ Open. 2012 Jun;2 doi: 10.1136/bmjopen-2012-000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee J, Mark RG. An investigation of patterns in hemodynamic data indicative of impending hypotension in intensive care. Biomed Eng Online. 2010 Oct;9 doi: 10.1186/1475-925X-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lau EC, et al. Use of electronic medical records (EMR) for oncology outcomes research: assessing the comparability of EMR information to patient registry and health claims data. Clin Epidemiol. 2011 Oct;3:259–272. doi: 10.2147/CLEP.S23690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Allasia G. Approximating potential integrals by cardinal basis interpolants on multivariate scattered data. Comput Math Appl. 2002 Feb-Mar;43:275–287. [Google Scholar]
- 122.Welch CA, et al. Evaluation of two-fold fully conditional specification multiple imputation for longitudinal electronic health record data. Statistics in Medicine. 2014 Sep;33:3725–3737. doi: 10.1002/sim.6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li Q, et al. Robust heart rate estimation from multiple asynchronous noisy sources using signal quality indices and a Kalman filter. Physiol Meas. 2008 Jan;29:15–32. doi: 10.1088/0967-3334/29/1/002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zong W, et al. Reduction of false arterial blood pressure alarms using signal quality assessment and relationships between the electrocardiogram and arterial blood pressure. Med Biol Eng Comput. 2004 Sep;42:698–706. doi: 10.1007/BF02347553. [DOI] [PubMed] [Google Scholar]
- 125.Silva I, et al. Signal Quality Estimation With Multichannel Adaptive Filtering in Intensive Care Settings. IEEE T Bio-Med Eng. 2012 Sep;59:2476–2485. doi: 10.1109/TBME.2012.2204882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Feldman JM, et al. Robust sensor fusion improves heart rate estimation: Clinical evaluation. J Clin Monitor. 1997 Nov;13:379–384. doi: 10.1023/a:1007476707284. [DOI] [PubMed] [Google Scholar]
- 127.Wartzek T, et al. Robust sensor fusion of unobtrusively measured heart rate. IEEE J Biomed Health Inform. 2014 Mar;18:654–660. doi: 10.1109/JBHI.2013.2274211. [DOI] [PubMed] [Google Scholar]
- 128.Labarère J, et al. How to derive and validate clinical prediction models for use in intensive care medicine. Intensive Care Medicine. 2014 Apr;40:513–527. doi: 10.1007/s00134-014-3227-6. [DOI] [PubMed] [Google Scholar]
- 129.Schafer JL. Multiple imputation: a primer. Statistical methods in medical research. 1999;8:3–15. doi: 10.1177/096228029900800102. [DOI] [PubMed] [Google Scholar]
- 130.Tian J, et al. Missing data analyses: a hybrid multiple imputation algorithm using Gray System Theory and entropy based on clustering. Applied Intelligence. 2014 Mar;40:376–388. [Google Scholar]
- 131.Hallgren KA, Witkiewitz K. Missing data in alcohol clinical trials: a comparison of methods. Alcohol Clin Exp Res. 2013 Dec;37:2152–2160. doi: 10.1111/acer.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Städler N, et al. Pattern Alternating Maximization Algorithm for Missing Data in High-Dimensional Problems. J Mach Learn Res. 2014 Jan;15:1903–1928. [Google Scholar]
- 133.Fuchs L, et al. ICU admission characteristics and mortality rates among elderly and very elderly patients. Intensive Care Medicine. 2012 Oct;38:1654–1661. doi: 10.1007/s00134-012-2629-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li Y, et al. A method for controlling complex confounding effects in the detection of adverse drug reactions using electronic health records. J Am Med Inform Assoc. 2014 Mar;21:308–314. doi: 10.1136/amiajnl-2013-001718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Andreao RV, et al. ECG signal analysis through hidden Markov models. IEEE T Bio-Med Eng. 2006 Aug;53:1541–1549. doi: 10.1109/TBME.2006.877103. [DOI] [PubMed] [Google Scholar]
- 136.de Lannoy G, et al. Weighted Conditional Random Fields for Supervised Interpatient Heartbeat Classification. IEEE T Bio-Med Eng. 2012 Jan;59:241–247. doi: 10.1109/TBME.2011.2171037. [DOI] [PubMed] [Google Scholar]
- 137.Klema J, et al. Sequential data mining: A comparative case study in development of atherosclerosis risk factors. IEEE T Syst Man Cy C. 2008 Jan;38:3–15. [Google Scholar]
- 138.Harpaz R, et al. Novel Data-Mining Methodologies for Adverse Drug Event Discovery and Analysis. Clin Pharmacol Ther. 2012 Jun;91:1010–1021. doi: 10.1038/clpt.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Harms SK, Deogun JS. Sequential association rule mining with time lags. J Intell Inf Syst. 2004 Jan;22:7–22. [Google Scholar]
- 140.Chen H, et al. Business Intelligence and Analytics: From Big Data to Big Impact. MIS Quarterly. 2012 Dec;36:1165–1188. [Google Scholar]
- 141.Ola O, Sedig K. The challenge of big data in public health: an opportunity for visual analytics. Online J Public Health Inform. 2014 Feb;5 doi: 10.5210/ojphi.v5i3.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Taylor RC. An overview of the Hadoop/MapReduce/HBase framework and its current applications in bioinformatics. BMC Bioinformatics. 2010 Dec;11(Suppl 12) doi: 10.1186/1471-2105-11-S12-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Biem A, et al. IBM infosphere streams for scalable, real-time, intelligent transportation services; Proceedings of the 2010 ACM SIGMOD International Conference on Management of Data; 2010. pp. 1093–1104. [Google Scholar]
- 144.Zaharia M, et al. Spark: cluster computing with working sets; Proceedings of the 2nd USENIX Conference on Hot Topics in Cloud Computing; 2010. [Google Scholar]
- 145.Demirkan H, Delen D. Leveraging the capabilities of service-oriented decision support systems: Putting analytics and big data in cloud. Decision Support Systems. 2013 May;55:412–421. [Google Scholar]
- 146.Illumina. BaseSpace: Genomics Cloud Computing. Available: https://basespace.illumina.com. [Google Scholar]
- 147.Afgan E, et al. Harnessing cloud computing with Galaxy Cloud. Nature Biotechnology. 2011 Nov;29:972–974. doi: 10.1038/nbt.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wright A, et al. Creating and sharing clinical decision support content with Web 2.0: Issues and examples. Journal of Biomedical Informatics. 2009;42:334–346. doi: 10.1016/j.jbi.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 149.T. C. G. Atlas. Available: http://cancergenome.nih.gov/
- 150.Kaddi CD, Wang MD. Models for Predicting Stage in Head and Neck Squamous Cell Carcinoma Using Proteomic Data; 2014 36th Annual International Conference of the Ieee Engineering in Medicine and Biology Society (Embc); 2014. pp. 5216–5219. [DOI] [PubMed] [Google Scholar]
- 151.Hoadley KA, et al. Multiplatform Analysis of 12 Cancer Types Reveals Molecular Classification within and across Tissues of Origin. Cell. 158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010 Jan 19;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.N. Cancer Genome Atlas. Comprehensive molecular portraits of human breast tumours. Nature. 2012 Oct 4;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.N. Cancer Genome Atlas Research. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012 Sep 27;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gottesman O, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013 Oct;15:761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Marsolo K, Spooner SA. Clinical genomics in the world of the electronic health record. Genet Med. 2013 Oct;15:786–791. doi: 10.1038/gim.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kannry JL, Williams MS. Integration of genomics into the electronic health record: mapping terra incognita. Genet Med. 2013 Jun;15:757–760. doi: 10.1038/gim.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ury AG. Storing and interpreting genomic information in widely deployed electronic health record systems. Genet Med. 2013 Aug;15:779–785. doi: 10.1038/gim.2013.111. [DOI] [PubMed] [Google Scholar]
- 159.Caligtan CA, Dykes PC. Electronic health records and personal health records. Semin Oncol Nurs. 2011 Aug;27:218–228. doi: 10.1016/j.soncn.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 160.Alterovitz G, et al. SMART on FHIR Genomics: facilitating standardized clinico-genomic apps. J Am Med Inform Assoc. 2015 Nov;22:1173–1178. doi: 10.1093/jamia/ocv045. [DOI] [PubMed] [Google Scholar]
- 161.NIH-BMIC. Available: https://www.nlm.nih.gov/NIHbmic/
- 162.WHO. Available: http://apps.who.int/gho/data/node.main.
- 163.NCHS. Available: http://www.healthindicators.gov/
- 164.Chen HC, et al. Business Intelligence and Analytics: From Big Data to Big Impact. MIS Quarterly. 2012 Dec;36:1165–1188. [Google Scholar]
- 165.Chen M, et al. Big Data: A Survey. Mobile Netw Appl. 2014 Apr;19:171–209. [Google Scholar]
- 166.Murff HJ, et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. Jama. 2011;306:848–855. doi: 10.1001/jama.2011.1204. [DOI] [PubMed] [Google Scholar]
- 167.McCarty CA, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC medical genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ricciardi L, et al. A national action plan to support consumer engagement via e-health. Health Affairs. 2013;32:376–384. doi: 10.1377/hlthaff.2012.1216. [DOI] [PubMed] [Google Scholar]
- 169.Keck MK, et al. Integrative analysis of Head and Neck Cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clinical Cancer Research. 2014 Dec 9; doi: 10.1158/1078-0432.CCR-14-2481. 2014. [DOI] [PubMed] [Google Scholar]
- 170.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nature Reviews Genetics. 2012 Apr;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Andreu-Perez J, et al. Big data for health. IEEE journal of biomedical and health informatics. 2015 Jul;19:1193–1208. doi: 10.1109/JBHI.2015.2450362. [DOI] [PubMed] [Google Scholar]
- 172.Cvach M. Monitor alarm fatigue: an integrative review. Biomed Instrum Technol. 2012 Jul;46:268–277. doi: 10.2345/0899-8205-46.4.268. [DOI] [PubMed] [Google Scholar]
- 173.Martis R, et al. Wavelet-based Machine Learning Techniques for ECG Signal Analysis. In: Dua S, Acharya UR, Dua P, editors. Machine Learning in Healthcare Informatics. Vol. 56. Berlin Heidelberg: Springer; 2014. pp. 25–45. [Google Scholar]