Figure 1.
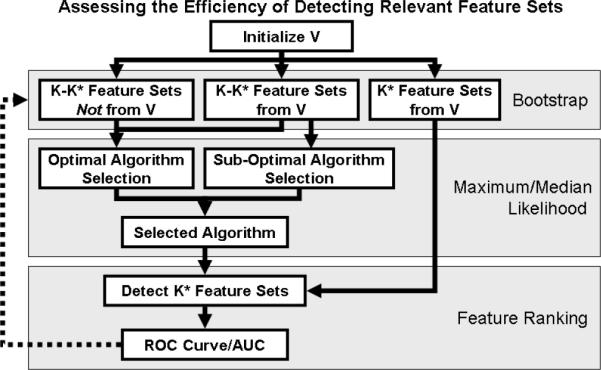
Quantifying the efficiency of detecting relevant feature sets. For clinical data, we define V as the set of K known differentially expressed feature sets. Using bootstrap cross validation, we partition V into K* and K-K* samples. K* is the number of unique samples after sampling from V K times with replacement. We optimize the ranking algorithm using K-K* feature sets and assess the algorithm's efficiency in detecting the remaining K* feature sets. For each of the three conditions— optimal metric selection, sub-optimal metric selection, and sub-optimal initial knowledge—we perform this bootstrap sampling 100 times in order to compute the significance of any differences between mean AUC values.
